Reconsidering the generation time hypothesis based on nuclear ribosomal ITS sequence comparisons in annual and perennial angiosperms
- PMID: 19113991
- PMCID: PMC2637270
- DOI: 10.1186/1471-2148-8-344
Reconsidering the generation time hypothesis based on nuclear ribosomal ITS sequence comparisons in annual and perennial angiosperms
Abstract
Background: Differences in plant annual/perennial habit are hypothesized to cause a generation time effect on divergence rates. Previous studies that compared rates of divergence for internal transcribed spacer (ITS1 and ITS2) sequences of nuclear ribosomal DNA (nrDNA) in angiosperms have reached contradictory conclusions about whether differences in generation times (or other life history features) are associated with divergence rate heterogeneity. We compared annual/perennial ITS divergence rates using published sequence data, employing sampling criteria to control for possible artifacts that might obscure any actual rate variation caused by annual/perennial differences.
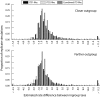
Results: Relative rate tests employing ITS sequences from 16 phylogenetically-independent annual/perennial species pairs rejected rate homogeneity in only a few comparisons, with annuals more frequently exhibiting faster substitution rates. Treating branch length differences categorically (annual faster or perennial faster regardless of magnitude) with a sign test often indicated an excess of annuals with faster substitution rates. Annuals showed an approximately 1.6-fold rate acceleration in nucleotide substitution models for ITS. Relative rates of three nuclear loci and two chloroplast regions for the annual Arabidopsis thaliana compared with two closely related Arabidopsis perennials indicated that divergence was faster for the annual. In contrast, A. thaliana ITS divergence rates were sometimes faster and sometimes slower than the perennial. In simulations, divergence rate differences of at least 3.5-fold were required to reject rate constancy in > 80 % of replicates using a nucleotide substitution model observed for the combination of ITS1 and ITS2. Simulations also showed that categorical treatment of branch length differences detected rate heterogeneity > 80% of the time with a 1.5-fold or greater rate difference.
Conclusion: Although rate homogeneity was not rejected in many comparisons, in cases of significant rate heterogeneity annuals frequently exhibited faster substitution rates. Our results suggest that annual taxa may exhibit a less than 2-fold rate acceleration at ITS. Since the rate difference is small and ITS lacks statistical power to reject rate homogeneity, further studies with greater power will be required to adequately test the hypothesis that annual and perennial plants have heterogeneous substitution rates. Arabidopsis sequence data suggest that relative rate tests based on multiple loci may be able to distinguish a weak acceleration in annual plants. The failure to detect rate heterogeneity with ITS in past studies may be largely a product of low statistical power.
Figures


Similar articles
-
Patterns of Substitution Rate Variation at Many Nuclear Loci in Two Species Trios in the Brassicaceae Partitioned with ANOVA.J Mol Evol. 2016 Oct;83(3-4):97-109. doi: 10.1007/s00239-016-9752-x. Epub 2016 Sep 3. J Mol Evol. 2016. PMID: 27592229
-
Genome-wide investigation reveals high evolutionary rates in annual model plants.BMC Plant Biol. 2010 Nov 9;10:242. doi: 10.1186/1471-2229-10-242. BMC Plant Biol. 2010. PMID: 21062446 Free PMC article.
-
Evolution of the mitochondrial rps3 intron in perennial and annual angiosperms and homology to nad5 intron 1.Mol Biol Evol. 1999 Apr;16(4):441-52. doi: 10.1093/oxfordjournals.molbev.a026126. Mol Biol Evol. 1999. PMID: 10331271
-
Characterization of angiosperm nrDNA polymorphism, paralogy, and pseudogenes.Mol Phylogenet Evol. 2003 Dec;29(3):435-55. doi: 10.1016/j.ympev.2003.08.021. Mol Phylogenet Evol. 2003. PMID: 14615185 Review.
-
Nuclear DNA amounts in angiosperms: targets, trends and tomorrow.Ann Bot. 2011 Mar;107(3):467-590. doi: 10.1093/aob/mcq258. Epub 2011 Jan 21. Ann Bot. 2011. PMID: 21257716 Free PMC article. Review.
Cited by
-
Patterns of Substitution Rate Variation at Many Nuclear Loci in Two Species Trios in the Brassicaceae Partitioned with ANOVA.J Mol Evol. 2016 Oct;83(3-4):97-109. doi: 10.1007/s00239-016-9752-x. Epub 2016 Sep 3. J Mol Evol. 2016. PMID: 27592229
-
Upper ocean oxygenation, evolution of RuBisCO and the Phanerozoic succession of phytoplankton.Free Radic Biol Med. 2019 Aug 20;140:295-304. doi: 10.1016/j.freeradbiomed.2019.05.006. Epub 2019 May 7. Free Radic Biol Med. 2019. PMID: 31075497 Free PMC article. Review.
-
Synteny analysis in Rosids with a walnut physical map reveals slow genome evolution in long-lived woody perennials.BMC Genomics. 2015 Sep 17;16(1):707. doi: 10.1186/s12864-015-1906-5. BMC Genomics. 2015. PMID: 26383694 Free PMC article.
-
Genome-wide investigation reveals high evolutionary rates in annual model plants.BMC Plant Biol. 2010 Nov 9;10:242. doi: 10.1186/1471-2229-10-242. BMC Plant Biol. 2010. PMID: 21062446 Free PMC article.
-
The uneven phylogeny and biogeography of Erodium (Geraniaceae): radiations in the Mediterranean and recent recurrent intercontinental colonization.Ann Bot. 2010 Dec;106(6):871-84. doi: 10.1093/aob/mcq184. Epub 2010 Sep 20. Ann Bot. 2010. PMID: 20858592 Free PMC article.
References
-
- Kimura M. The neutral theory of molecular evolution. New York: Cambridge University Press; 1983.
-
- Gillespie JH. The causes of molecular evolution. New York: Oxford University Press; 1991.
-
- Ohta T. The nearly neutral theory of molecular evolution. Annu Rev Syst Ecol. 1992;23:263–286. doi: 10.1146/annurev.es.23.110192.001403. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

