SR proteins and the nonsense-mediated decay mechanism are involved in human GLB1 gene alternative splicing
- PMID: 19114006
- PMCID: PMC2631023
- DOI: 10.1186/1756-0500-1-137
SR proteins and the nonsense-mediated decay mechanism are involved in human GLB1 gene alternative splicing
Abstract
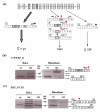
Background: The human GLB1 gene is known to give rise to two alternatively spliced mRNAs, which encode two different proteins: lysosomal beta-galactosidase (beta-gal) and elastin-binding protein (EBP). The beta-gal transcript includes the 16 exons of the GLB1 gene. In the EBP transcript, exons 3, 4 and 6 are skipped, while exon 5 has a different reading frame. However, little is known on how this alternative splicing is regulated.
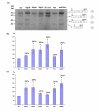
Findings: Cycloheximide treatment of HeLa cells and human fibroblasts revealed the presence of new transcripts that are otherwise degraded by nonsense-mediated decay (NMD). A minigene carrying the exons involved in the alternative splicing of GLB1 was constructed. Improving the acceptor-site scores of exons 3 or 4 increased the relative inclusion of these exons, but did not stop them being skipped in some transcripts. Overexpression of different SR proteins altered the relative proportion of the different transcripts produced by the minigene, indicating a possible mechanism for the regulation of the alternative splicing of GLB1. Finally, a comparison of this gene among different species was performed.
Conclusion: In the processing of the GLB1 RNA several transcripts are generated, but only those with a correct reading frame are not degraded. The differential inclusion/exclusion of exons could be partially explained by the relatively weak splice sites in the exons involved. Different SR proteins have an effect on the process of skipping of these exons, at least in the minigene conditions, indicating a possible mechanism for the regulation of the alternative splicing of the GLB1 gene.
Figures


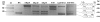
References
LinkOut - more resources
Full Text Sources
Research Materials

