Archaeal ApbC/Nbp35 homologs function as iron-sulfur cluster carrier proteins
- PMID: 19114487
- PMCID: PMC2648223
- DOI: 10.1128/JB.01469-08
Archaeal ApbC/Nbp35 homologs function as iron-sulfur cluster carrier proteins
Abstract
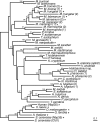
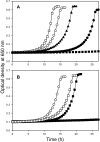
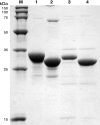
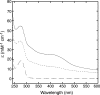
Iron-sulfur clusters may have been the earliest catalytic cofactors on earth, and most modern organisms use them extensively. Although members of the Archaea produce numerous iron-sulfur proteins, the major cluster assembly proteins found in the Bacteria and Eukarya are not universally conserved in archaea. Free-living archaea do have homologs of the bacterial apbC and eukaryotic NBP35 genes that encode iron-sulfur cluster carrier proteins. This study exploits the genetic system of Salmonella enterica to examine the in vivo functionality of apbC/NBP35 homologs from three archaea: Methanococcus maripaludis, Methanocaldococcus jannaschii, and Sulfolobus solfataricus. All three archaeal homologs could correct the tricarballylate growth defect of an S. enterica apbC mutant. Additional genetic studies showed that the conserved Walker box serine and the Cys-X-X-Cys motif of the M. maripaludis MMP0704 protein were both required for function in vivo but that the amino-terminal ferredoxin domain was not. MMP0704 protein and an MMP0704 variant protein missing the N-terminal ferredoxin domain were purified, and the Fe-S clusters were chemically reconstituted. Both proteins bound equimolar concentrations of Fe and S and had UV-visible spectra similar to those of known [4Fe-4S] cluster-containing proteins. This family of dimeric iron-sulfur carrier proteins evolved before the archaeal and eukaryal lineages diverged, representing an ancient mode of cluster assembly.
Figures

References
-
- Agar, J. N., P. Yuvaniyama, R. F. Jack, V. L. Cash, A. D. Smith, D. R. Dean, and M. K. Johnson. 2000. Modular organization and identification of a mononuclear iron-binding site within the NifU protein. J. Biol. Inorg. Chem. 5167-177. - PubMed
-
- Bartolomé, B., Y. Jubete, E. Martínez, and F. de la Cruz. 1991. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene 10275-78. - PubMed
-
- Beinert, H. 1983. Semi-micro methods for analysis of labile sulfide and of labile sulfide plus sulfane sulfur in unusually stable iron-sulfur proteins. Anal. Biochem. 131373-378. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

