Molecular diversity of a North Carolina wastewater treatment plant as revealed by pyrosequencing
- PMID: 19114525
- PMCID: PMC2655459
- DOI: 10.1128/AEM.01210-08
Molecular diversity of a North Carolina wastewater treatment plant as revealed by pyrosequencing
Abstract
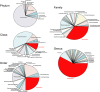
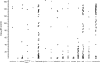
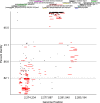
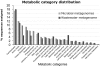
We report the results of pyrosequencing of DNA collected from the activated sludge basin of a wastewater treatment plant in Charlotte, NC. Using the 454-FLX technology, we generated 378,601 sequences with an average read length of 250.4 bp. Running the 454 assembly algorithm over our sequences yielded very poor assembly, with only 0.3% of our sequences participating in assembly of significant contigs. Of the 117 contigs greater than 500 bp long that were assembled, the most common annotations were to transposases and hypothetical proteins. Comparing our sequences to known microbial genomes showed nonspecific recruitment, indicating that previously described taxa are only distantly related to the most abundant microbes in this treatment plant. A comparison of proteins generated by translating our sequence set to translations of other sequenced microbiomes shows a distinct metabolic profile for activated sludge with high counts for genes involved in metabolism of aromatic compounds and low counts for genes involved in photosynthesis. Taken together, these data document the substantial levels of microbial diversity within activated sludge and further establish the great utility of pyrosequencing for investigating diversity in complex ecosystems.
Figures



References
-
- Amann, R., H. Lemmer, and M. Wagner. 1998. Monitoring the community structure of wastewater treatment plants: a comparison of old and new techniques. FEMS Microbiol. Ecol. 25:205-215.
-
- Aziz, R. K., D. Bartels, A. A. Best, M. DeJongh, T. Disz, R. A. Edwards, K. Formsma, S. Gerdes, E. M. Glass, M. Kubal, F. Meyer, G. J. Olsen, R. Olson, A. L. Osterman, R. A. Overbeek, L. K. McNeil, D. Paarmann, T. Paczian, B. Parrello, G. D. Pusch, C. Reich, R. Stevens, O. Vassieva, V. Vonstein, A. Wilke, and O. Zagnitko. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. - PMC - PubMed
-
- Beline, F., J. Martinez, C. Marol, and G. Guiraud. 2001. Application of the 15N technique to determine the contributions of nitrification and denitrification to the flux of nitrous oxide from aerated pig slurry. Water Res. 65:2774-2778. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

