Ikaros and GATA-1 combinatorial effect is required for silencing of human gamma-globin genes
- PMID: 19114560
- PMCID: PMC2648246
- DOI: 10.1128/MCB.01523-08
Ikaros and GATA-1 combinatorial effect is required for silencing of human gamma-globin genes
Abstract
During development and erythropoiesis, globin gene expression is finely modulated through an important network of transcription factors and chromatin modifying activities. In this report we provide in vivo evidence that endogenous Ikaros is recruited to the human beta-globin locus and targets the histone deacetylase HDAC1 and the chromatin remodeling protein Mi-2 to the human gamma-gene promoters, thereby contributing to gamma-globin gene silencing at the time of the gamma- to beta-globin gene transcriptional switch. We show for the first time that Ikaros interacts with GATA-1 and enhances the binding of the latter to different regulatory regions across the locus. Consistent with these results, we show that the combinatorial effect of Ikaros and GATA-1 impairs close proximity between the locus control region and the human gamma-globin genes. Since the absence of Ikaros also affects GATA-1 recruitment to GATA-2 promoter, we propose that the combinatorial effect of Ikaros and GATA-1 is not restricted to globin gene regulation.
Figures




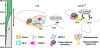
References
-
- Avitahl, N., S. Winandy, C. Friedrich, B. Jones, Y. Ge, and K. Georgopoulos. 1999. Ikaros sets thresholds for T-cell activation and regulates chromosome propagation. Immunity 10333-343. - PubMed
-
- Cantor, A. B., and S. H. Orkin. 2005. Coregulation of GATA factors by the Friend of GATA (FOG) family of multitype zinc finger proteins. Semin. Cell Dev. Biol. 16117-128. - PubMed
-
- Cantor, A. B., and S. H. Orkin. 2002. Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene 213368-3376. - PubMed
-
- Carter, D., L. Chakalova, C. S. Osborne, Y. F. Dai, and P. Fraser. 2002. Long-range chromatin regulatory interactions in vivo. Nat. Genet. 32623-626. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
