A regulatory interplay between miR-27a and Runx1 during megakaryopoiesis
- PMID: 19114653
- PMCID: PMC2612036
- DOI: 10.1073/pnas.0811466106
A regulatory interplay between miR-27a and Runx1 during megakaryopoiesis
Abstract
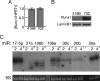
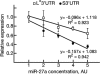
The transcription factor Runx1 is a key regulator of definitive hematopoiesis in the embryo and the adult. Lineage-specific expression of Runx1 involves transcription and post-transcription control through usage of alternative promoters and diverse 3'UTR isoforms, respectively. We identified and mapped microRNA (miR) binding sites on Runx1 3'UTR and show that miR-27a, miR-9, miR-18a, miR-30c, and miR-199a* bind and post-transcriptionally attenuate expression of Runx1. miR-27a impacts on both the shortest (0.15 kb) and longest (3.8 kb) 3'UTRs and, along with additional miRs, might contribute to translation attenuation of Runx1 mRNA in the myeloid cell line 416B. Whereas levels of Runx1 mRNA in 416B and the B cell line 70Z were similar, the protein levels were not. Large amounts of Runx1 protein were found in 70Z cells, whereas only minute amounts of Runx1 protein were made in 416B cells and overexpression of Runx1 in 416B induced terminal differentiation associated with megakaryocytic markers. Induction of megakaryocytic differentiation in K562 cells by 12-o-tetradecanoylphorbol-13-acetate markedly increased miR-27a expression, concomitantly with binding of Runx1 to miR-27a regulatory region. The data indicate that miR-27a plays a regulatory role in megakaryocytic differentiation by attenuating Runx1 expression, and that, during megakaryopoiesis, Runx1 and miR-27a are engaged in a feedback loop involving positive regulation of miR-27a expression by Runx1.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Cameron ER, Neil JC. The Runx genes: lineage-specific oncogenes and tumor suppressors. Oncogene. 2004;23:4308–4314. - PubMed
-
- De Brujin MF, Speck NA. Core-binding factors in hematopoiesis and immune function. Oncogene. 2004;23:4238–4248. - PubMed
-
- Levanon D, Groner Y. Structure and regulated expression of mammalian RUNX genes. Oncogene. 2004;23:4211–4219. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

