Function and assembly of a chromatin-associated RNase P that is required for efficient transcription by RNA polymerase I
- PMID: 19115013
- PMCID: PMC2605565
- DOI: 10.1371/journal.pone.0004072
Function and assembly of a chromatin-associated RNase P that is required for efficient transcription by RNA polymerase I
Abstract
Background: Human RNase P has been initially described as a tRNA processing enzyme, consisting of H1 RNA and at least ten distinct protein subunits. Recent findings, however, indicate that this catalytic ribonucleoprotein is also required for transcription of small noncoding RNA genes by RNA polymerase III (Pol III). Notably, subunits of human RNase P are localized in the nucleolus, thus raising the possibility that this ribonucleoprotein complex is implicated in transcription of rRNA genes by Pol I.
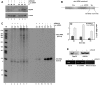
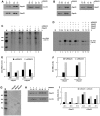
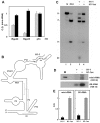
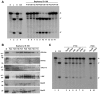
Methodology/principal findings: By using biochemical and reverse genetic means we show here that human RNase P is required for efficient transcription of rDNA by Pol I. Thus, inactivation of RNase P by targeting its protein subunits for destruction by RNA interference or its H1 RNA moiety for specific cleavage causes marked reduction in transcription of rDNA by Pol I. However, RNase P restores Pol I transcription in a defined reconstitution system. Nuclear run on assays reveal that inactivation of RNase P reduces the level of nascent transcription by Pol I, and more considerably that of Pol III. Moreover, RNase P copurifies and associates with components of Pol I and its transcription factors and binds to chromatin of the promoter and coding region of rDNA. Strikingly, RNase P detaches from transcriptionally inactive rDNA in mitosis and reassociates with it at G1 phase through a dynamic and stepwise assembly process that is correlated with renewal of transcription.
Conclusions/significance: Our findings reveal that RNase P activates transcription of rDNA by Pol I through a novel assembly process and that this catalytic ribonucleoprotein determines the transcription output of Pol I and Pol III, two functionally coordinated transcription machineries.
Conflict of interest statement
Figures



References
-
- Wassarman KM. RNA regulators of transcription. Nat Struct Mol Biol. 2004;11:803–804. - PubMed
-
- Storz G, Altuvia S, Wassarman KM. An abundance of RNA regulators. Annu Rev Biochem. 2005;74:199–217. - PubMed
-
- Mayer C, Schmitz KM, Li J, Grummt I, Santoro R. Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol Cell. 2006;22:351–361. - PubMed
-
- Kettenberger H, Eisenführ A, Brueckner F, Theis M, Famulok M, et al. Structure of an RNA polymerase II-RNA inhibitor complex elucidates transcription regulation by noncoding RNAs. Nat Struct Mol Biol. 2006;13:44–48. - PubMed
-
- Goodrich JA, Kugel JF. Non-coding-RNA regulators of RNA polymerase II transcription. Nat Rev Mol Cell Biol. 2006;7:612–616. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

