Ras acylation, compartmentalization and signaling nanoclusters (Review)
- PMID: 19115142
- PMCID: PMC2782584
- DOI: 10.1080/09687680802649582
Ras acylation, compartmentalization and signaling nanoclusters (Review)
Abstract
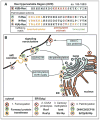
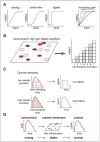
Ras proteins have become paradigms for isoform- and compartment-specific signaling. Recent work has shown that Ras isoforms are differentially distributed within cell surface signaling nanoclusters and on endomembranous compartments. The critical feature regulating Ras protein localization and isoform-specific functions is the C-terminal hypervariable region (HVR). In this review we discuss the differential post-translational modifications and reversible targeting functions of Ras isoform HVR motifs. We describe how compartmentalized Ras signaling has specific functional consequences and how cell surface signaling nanoclusters generate precise signaling outputs.
Figures

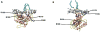
References
-
- Esteban LM, Vicario-Abejon C, Fernandez-Salguero P, Fernandez-Medarde A, Swaminathan N, Yienger K, Lopez E, Malumbres M, McKay R, Ward JM, Pellicer A, Santos E. Targeted genomic disruption of H-ras and N-ras, individually or in combination, reveals the dispensability of both loci for mouse growth and development. Mol Cell Biol. 2001;21:1444–1452. - PMC - PubMed
-
- The data was obtained from the Wellcome Trust Sanger Institute Cancer Genome Project web site. http://www.sanger.ac.uk/genetics/CGP.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
