Genome based cell population heterogeneity promotes tumorigenicity: the evolutionary mechanism of cancer
- PMID: 19115235
- PMCID: PMC2778062
- DOI: 10.1002/jcp.21663
Genome based cell population heterogeneity promotes tumorigenicity: the evolutionary mechanism of cancer
Abstract
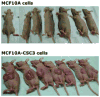
Cancer progression represents an evolutionary process where overall genome level changes reflect system instability and serve as a driving force for evolving new systems. To illustrate this principle it must be demonstrated that karyotypic heterogeneity (population diversity) directly contributes to tumorigenicity. Five well characterized in vitro tumor progression models representing various types of cancers were selected for such an analysis. The tumorigenicity of each model has been linked to different molecular pathways, and there is no common molecular mechanism shared among them. According to our hypothesis that genome level heterogeneity is a key to cancer evolution, we expect to reveal that the common link of tumorigenicity between these diverse models is elevated genome diversity. Spectral karyotyping (SKY) was used to compare the degree of karyotypic heterogeneity displayed in various sublines of these five models. The cell population diversity was determined by scoring type and frequencies of clonal and non-clonal chromosome aberrations (CCAs and NCCAs). The tumorigenicity of these models has been separately analyzed. As expected, the highest level of NCCAs was detected coupled with the strongest tumorigenicity among all models analyzed. The karyotypic heterogeneity of both benign hyperplastic lesions and premalignant dysplastic tissues were further analyzed to support this conclusion. This common link between elevated NCCAs and increased tumorigenicity suggests an evolutionary causative relationship between system instability, population diversity, and cancer evolution. This study reconciles the difference between evolutionary and molecular mechanisms of cancer and suggests that NCCAs can serve as a biomarker to monitor the probability of cancer progression.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

