Identification of a novel point mutation in ENT1 that confers resistance to Ara-C in human T cell leukemia CCRF-CEM cells
- PMID: 19116148
- PMCID: PMC2647365
- DOI: 10.1016/j.febslet.2008.12.041
Identification of a novel point mutation in ENT1 that confers resistance to Ara-C in human T cell leukemia CCRF-CEM cells
Abstract
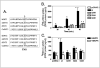
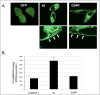
The genetic basis for the Ara-C resistance of CCRF-CEM Ara-C/8C leukemia cells was investigated. DNA sequencing revealed that these cells expressed an equilibrative nucleoside transporter 1 (ENT1) with a single missense mutation resulting in glycine to arginine replacement (G24R). To test the importance of this residue, additional G24 mutants were created and examined for [3H]-uridine and [3H]-Ara-C uptake. Both a G24E and G24A mutant showed reduced ENT1-dependent activity. An EGFP-tagged G24R ENT1 displayed plasma membrane localization even though it was unable to bind [3H]-NBMPR, an ENT1-specific inhibitor. These results define G24 as critical amino acid for ENT1 nucleoside uptake and suggest that mutations in TM1 may provide a mechanism for Ara-C resistance in CCRF-CEM Ara-C/8C cells.
Figures

Similar articles
-
Gene-expression profiling reveals down-regulation of equilibrative nucleoside transporter 1 (ENT1) in Ara-C-resistant CCRF-CEM-derived cells.J Biochem. 2004 Nov;136(5):733-40. doi: 10.1093/jb/mvh180. J Biochem. 2004. PMID: 15632314
-
Role of equilibrative nucleoside transporter 1 (ENT1) in the disposition of cytarabine in mice.Pharmacol Res Perspect. 2019 Dec 2;7(6):e00534. doi: 10.1002/prp2.534. eCollection 2019 Dec. Pharmacol Res Perspect. 2019. PMID: 31832201 Free PMC article.
-
Two distinct molecular mechanisms underlying cytarabine resistance in human leukemic cells.Cancer Res. 2008 Apr 1;68(7):2349-57. doi: 10.1158/0008-5472.CAN-07-5528. Cancer Res. 2008. PMID: 18381442
-
FLT3-ITD induces ara-C resistance in myeloid leukemic cells through the repression of the ENT1 expression.Biochem Biophys Res Commun. 2009 Dec 18;390(3):1001-6. doi: 10.1016/j.bbrc.2009.10.094. Epub 2009 Oct 22. Biochem Biophys Res Commun. 2009. PMID: 19853583
-
Genetic factors influencing cytarabine therapy.Pharmacogenomics. 2009 Oct;10(10):1657-74. doi: 10.2217/pgs.09.118. Pharmacogenomics. 2009. PMID: 19842938 Free PMC article. Review.
Cited by
-
Nucleoside transporters and immunosuppressive adenosine signaling in the tumor microenvironment: Potential therapeutic opportunities.Pharmacol Ther. 2022 Dec;240:108300. doi: 10.1016/j.pharmthera.2022.108300. Epub 2022 Oct 22. Pharmacol Ther. 2022. PMID: 36283452 Free PMC article. Review.
-
Response and Toxicity to Cytarabine Therapy in Leukemia and Lymphoma: From Dose Puzzle to Pharmacogenomic Biomarkers.Cancers (Basel). 2021 Feb 25;13(5):966. doi: 10.3390/cancers13050966. Cancers (Basel). 2021. PMID: 33669053 Free PMC article. Review.
-
Inhibition of the equilibrative nucleoside transporter 1 and activation of A2A adenosine receptors by 8-(4-chlorophenylthio)-modified cAMP analogs and their hydrolytic products.J Biol Chem. 2009 Nov 20;284(47):32256-63. doi: 10.1074/jbc.M109.056622. Epub 2009 Sep 29. J Biol Chem. 2009. PMID: 19801629 Free PMC article.
-
Equilibrative nucleoside transporter 1 expression in primary human hepatocytes is highly variable and determines uptake of ribavirin.Antivir Chem Chemother. 2017 Apr;25(1):2-10. doi: 10.1177/2040206616686894. Epub 2017 Jan 1. Antivir Chem Chemother. 2017. PMID: 28417642 Free PMC article.
-
[Functional study of hENT1 on SKM-1 cell resistance to decitabine].Zhonghua Xue Ye Xue Za Zhi. 2015 May;36(5):408-12. doi: 10.3760/cma.j.issn.0253-2727.2015.05.012. Zhonghua Xue Ye Xue Za Zhi. 2015. PMID: 26031529 Free PMC article. Chinese.
References
-
- Cripe LD. Adult acute leukemia. Curr Probl Cancer. 1997;21(1):1–64. - PubMed
-
- Ullman B, Coons T, Rockwell S, McCartan K. Genetic analysis of 2′,3′-dideoxycytidine incorporation into cultured human T lymphoblasts. J Biol Chem. 1988;263:12391–96. - PubMed
-
- Ullman B. Dideoxycytidine metabolism in wild type and mutant CEM cells deficient in nucleoside transport or deoxycytidine kinase. Adv Exp Med Biol. 1989;253B:415–20. - PubMed
-
- Baldwin SA, Beal PR, Yao SY, King AE, Cass CE, Young JD. The equilibrative nucleoside transporter family, SLC29. Pflugers Arch. 2004;447:735–43. - PubMed
-
- Molina-Arcas M, Marce S, Villamor N, Huber-Ruano I, Casado FJ, Bellosillo B, Montserrat E, Gil J, Colomer D, Pastor-Anglada M. Equilibrative nucleoside transporter-2 (hENT2) protein expression correlates with ex vivo sensitivity to fludarabine in chronic lymphocytic leukemia (CLL) cells. Leukemia. 2005;19:64–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

