3D imaging of mammalian cells with ion-abrasion scanning electron microscopy
- PMID: 19116171
- PMCID: PMC4804765
- DOI: 10.1016/j.jsb.2008.11.005
3D imaging of mammalian cells with ion-abrasion scanning electron microscopy
Abstract
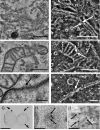
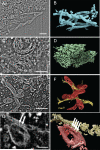
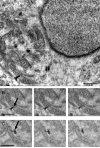
Understanding the hierarchical organization of molecules and organelles within the interior of large eukaryotic cells is a challenge of fundamental interest in cell biology. We are using ion-abrasion scanning electron microscopy (IA-SEM) to visualize this hierarchical organization in an approach that combines focused ion-beam milling with scanning electron microscopy. Here, we extend our previous studies on imaging yeast cells to image subcellular architecture in human melanoma cells and melanocytes at resolutions as high as approximately 6 and approximately 20 nm in the directions parallel and perpendicular, respectively, to the direction of ion-beam milling. The 3D images demonstrate the striking spatial relationships between specific organelles such as mitochondria and membranes of the endoplasmic reticulum, and the distribution of unique cellular components such as melanosomes. We also show that 10nm-sized gold particles and quantum dot particles with 7 nm-sized cores can be detected in single cross-sectional images. IA-SEM is thus a useful tool for imaging large mammalian cells in their entirety at resolutions in the nanometer range.
Figures


References
-
- Balaban RS. Modeling mitochondrial function. Am. J. Physiol. Cell Physiol. 2006;291:C1107–C1113. - PubMed
-
- Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313 - PubMed
-
- Birkedal R, Shiels HA, Vendelin M. Three-dimensional mitochondrial arrangement in ventricular myocytes: from chaos to order. Am. J. Physiol. Cell Physiol. 2006;291:C1148–C1158. - PubMed
-
- Dell'Angelica EC. The building BLOC(k)s of lysosomes and related organelles. Curr. Opin. Cell Biol. 2004;16 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

