Structure and activity of the human growth hormone receptor (hGHR) gene V2 promoter
- PMID: 19116246
- PMCID: PMC5428157
- DOI: 10.1210/me.2008-0188
Structure and activity of the human growth hormone receptor (hGHR) gene V2 promoter
Abstract
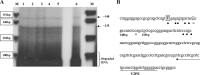
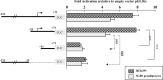
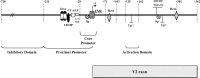
Human GH (hGH) has important effects on growth as well as carbohydrate, fat, and protein metabolism. These actions require the presence of normal levels of a functional hGH receptor (hGHR) on the surface of target cells. hGHR gene expression is characterized by the use of several 5'-noncoding exons and alternative splicing, resulting in the generation of multiple mRNA isoforms. The hGHR V2 transcript is predominant in most tissues, including human fat. However, factors regulating its ubiquitous expression have remained unidentified. The present study was aimed at characterizing the mechanisms regulating hGHR V2 transcription. Two major V2 transcriptional start sites were identified by primer extension assays. The V2 proximal promoter is TATA-less, with several characteristics of a housekeeping gene promoter. Transient transfection analyses of 2.6 kb of the 5'-flanking region of V2 confirmed its promoter activity in multiple primate cell lines. Similar promoter activity patterns were observed in human SGBS preadipocytes and mature adipocytes but with much higher V2 promoter activity in mature adipocytes, suggesting that changes in the availability of specific factors during adipocyte differentiation play a role in V2 promoter regulation. Serial deletion and mutation analyses revealed that transcription of hGHR V2 in different cell types, including adipocytes, is determined by a core promoter and distinct inhibitory and activation domains in the 5'-promoter region as well as within the V2 exon. Our data suggest that V2 transcription is the result of a complex interplay involving multiple factors, to ensure appropriate expression of hGHR in different hGH target cells.
Figures





Similar articles
-
Transcriptional regulation of the human growth hormone receptor (hGHR) gene V2 promoter by transcriptional activators and repressor.Mol Endocrinol. 2009 Mar;23(3):373-87. doi: 10.1210/me.2008-0190. Epub 2008 Dec 30. Mol Endocrinol. 2009. PMID: 19116245 Free PMC article.
-
Characterization of growth hormone receptor messenger ribonucleic acid variants in human adipocytes.J Clin Endocrinol Metab. 2006 May;91(5):1901-8. doi: 10.1210/jc.2005-1802. Epub 2006 Feb 21. J Clin Endocrinol Metab. 2006. PMID: 16492697
-
Expression of the hepatic specific V1 messenger ribonucleic acid of the human growth hormone receptor gene is regulated by hepatic nuclear factor (HNF)-4alpha2 and HNF-4alpha8.Mol Endocrinol. 2008 Feb;22(2):485-500. doi: 10.1210/me.2007-0387. Epub 2007 Nov 8. Mol Endocrinol. 2008. PMID: 17991764 Free PMC article.
-
Alu elements in human growth hormone receptor gene 5' untranslated region exons.J Mol Endocrinol. 2001 Dec;27(3):357-66. doi: 10.1677/jme.0.0270357. J Mol Endocrinol. 2001. PMID: 11719288
-
Organization and evolution of the human growth hormone receptor gene 5'-flanking region.Endocrinology. 2001 May;142(5):1923-34. doi: 10.1210/endo.142.5.8170. Endocrinology. 2001. PMID: 11316758
Cited by
-
Transcriptional regulation of the human growth hormone receptor (hGHR) gene V2 promoter by transcriptional activators and repressor.Mol Endocrinol. 2009 Mar;23(3):373-87. doi: 10.1210/me.2008-0190. Epub 2008 Dec 30. Mol Endocrinol. 2009. PMID: 19116245 Free PMC article.
References
-
- Perry JK, Emerald BS, Mertani HC, Lobie PE2006. The oncogenic potential of growth hormone. Growth Horm IGF Res 16:277–289 - PubMed
-
- Waters MJ, Shang CA, Behncken SN, Tam SP, Li H, Shen B, Lobie PE1999. Growth hormone as a cytokine. Clin Exp Pharmacol Physiol 26:760–764 - PubMed
-
- Wabitsch M, Hauner H, Heinze E, Teller W1994. In vitro effects of growth hormone in adipose tissue. Acta Paediatr Suppl 406:48–53 - PubMed
-
- Wabitsch M, Hauner H, Heinze E, Teller WM1995. The role of growth hormone/insulin-like growth factors in adipocyte differentiation. Metabolism 44:45–49 - PubMed
-
- Wabitsch M, Braun S, Hauner H, Heinze E, Ilondo MM, Shymko R, De Meyts P, Teller WM1996. Mitogenic and antiadipogenic properties of human growth hormone in differentiating human adipocyte precursor cells in primary culture. Pediatr Res 40:450–456 - PubMed

