Identification of direct residue contacts in protein-protein interaction by message passing
- PMID: 19116270
- PMCID: PMC2629192
- DOI: 10.1073/pnas.0805923106
Identification of direct residue contacts in protein-protein interaction by message passing
Abstract
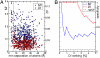
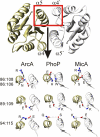
Understanding the molecular determinants of specificity in protein-protein interaction is an outstanding challenge of postgenome biology. The availability of large protein databases generated from sequences of hundreds of bacterial genomes enables various statistical approaches to this problem. In this context covariance-based methods have been used to identify correlation between amino acid positions in interacting proteins. However, these methods have an important shortcoming, in that they cannot distinguish between directly and indirectly correlated residues. We developed a method that combines covariance analysis with global inference analysis, adopted from use in statistical physics. Applied to a set of >2,500 representatives of the bacterial two-component signal transduction system, the combination of covariance with global inference successfully and robustly identified residue pairs that are proximal in space without resorting to ad hoc tuning parameters, both for heterointeractions between sensor kinase (SK) and response regulator (RR) proteins and for homointeractions between RR proteins. The spectacular success of this approach illustrates the effectiveness of the global inference approach in identifying direct interaction based on sequence information alone. We expect this method to be applicable soon to interaction surfaces between proteins present in only 1 copy per genome as the number of sequenced genomes continues to expand. Use of this method could significantly increase the potential targets for therapeutic intervention, shed light on the mechanism of protein-protein interaction, and establish the foundation for the accurate prediction of interacting protein partners.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Cusick ME, Klitgord N, Vidal M, Hill DE. Interactome: Gateway into systems biology. Hum Mol Genet. 2005;14((Spec No 2)):R171–R181. - PubMed
-
- Kortemme T, Baker D. Computational design of protein-protein interactions. Curr Opin Chem Biol. 2004;8:91–97. - PubMed
-
- Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450:1001–1009. - PubMed
-
- Suel GM, Lockless SW, Wall MA, Ranganathan R. Evolutionarily conserved networks of residues mediate allosteric communication in proteins. Nat Struct Biol. 2003;10:59–69. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

