Galectin-1 induces reversible phosphatidylserine exposure at the plasma membrane
- PMID: 19116313
- PMCID: PMC2649277
- DOI: 10.1091/mbc.e08-07-0786
Galectin-1 induces reversible phosphatidylserine exposure at the plasma membrane
Abstract
Cells normally undergo physiological turnover through the induction of apoptosis and phagocytic removal, partly through exposure of cell surface phosphatidylserine (PS). In contrast, neutrophils appear to possess apoptosis-independent mechanisms of removal. Here we show that Galectin-1 (Gal-1) induces PS exposure independent of alterations in mitochondrial potential, caspase activation, or cell death. Furthermore, Gal-1-induced PS exposure reverts after Gal-1 removal without altering cell viability. Gal-1-induced PS exposure is uniquely microdomain restricted, yet cells exposing PS do not display evident alterations in membrane morphology nor do they exhibit bleb formation, typically seen in apoptotic cells. Long-term exposure to Gal-1 prolongs PS exposure with no alteration in cell cycle progression or cell growth. These results demonstrate that Gal-1-induced PS exposure and subsequent phagocytic removal of living cells represents a new paradigm in cellular turnover.
Figures

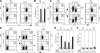
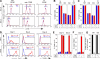
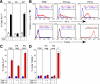
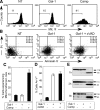




References
-
- Adams L., Scott G. K., Weinberg C. S. Biphasic modulation of cell growth by recombinant human galectin-1. Biochim. Biophys. Acta. 1996;1312:137–144. - PubMed
-
- Antia R., Ganusov V. V., Ahmed R. The role of models in understanding CD8+ T-cell memory. Nat. Rev. Immunol. 2005;5:101–111. - PubMed
-
- Barber D. L., Wherry E. J., Ahmed R. Cutting edge: rapid in vivo killing by memory CD8 T cells. J. Immunol. 2003;171:27–31. - PubMed
-
- Carlow D. A., Williams M. J., Ziltener H. J. Modulation of O-glycans and N-glycans on murine CD8 T cells fails to alter annexin V ligand induction by galectin 1. J. Immunol. 2003;171:5100–5106. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

