Molecular profiling of T-helper immune genes during dengue virus infection
- PMID: 19117515
- PMCID: PMC2628356
- DOI: 10.1186/1743-422X-5-165
Molecular profiling of T-helper immune genes during dengue virus infection
Abstract
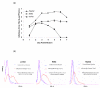
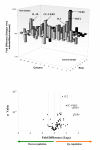
In this study, we provide a comprehensive molecular profiling of the involvement of T- helper (Th) genes during dengue virus infection of different cell types. The Th gene profiles of three human cell types (monocytes, T-cells and hepatocytes) were analyzed simultaneously via array-based RT-PCR upon infection with dengue virus. Differential regulation of 41 Th genes was identified and of which 20 of those genes may contribute to immuno-pathogenesis of dengue virus infection by regulating inflammation, thrombocytopenia and vascular permeability. Among the strongly up-regulated genes were the RANTES, CC-CKR3, IRF4, CLEC2C, IL-6 and TLR6, which are potent inducer of inflammation and vascular permeability. Profiling genes obtained from this study may serve as potential biomarkers and the modulation of Th immune responses during dengue virus infection has important implications in disease outcome.
Figures


References
-
- Dengue haemorrhagic fever: early recognition, diagnosis and hospital management–an audiovisual guide for health-care workers responding to outbreaks. Wkly Epidemiol Rec. 2006;81:362–363. - PubMed
-
- Warke RV, Xhaja K, Martin KJ, Fournier MF, Shaw SK, Brizuela N, de Bosch N, Lapointe D, Ennis FA, Rothman AL, Bosch I. Dengue virus induces novel changes in gene expression of human umbilical vein endothelial cells. J Virol. 2003;77:11822–11832. doi: 10.1128/JVI.77.21.11822-11832.2003. - DOI - PMC - PubMed
-
- Simmons CP, Popper S, Dolocek C, Chau TN, Griffiths M, Dung NT, Long TH, Hoang DM, Chau NV, Thao le TT, et al. Patterns of host genome-wide gene transcript abundance in the peripheral blood of patients with acute dengue hemorrhagic fever. J Infect Dis. 2007;195:1097–1107. doi: 10.1086/512162. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

