Detergent resistant membrane-associated IDE in brain tissue and cultured cells: Relevance to Abeta and insulin degradation
- PMID: 19117523
- PMCID: PMC2648957
- DOI: 10.1186/1750-1326-3-22
Detergent resistant membrane-associated IDE in brain tissue and cultured cells: Relevance to Abeta and insulin degradation
Abstract
Background: Insulin degrading enzyme (IDE) is implicated in the regulation of amyloid beta (Abeta) steady-state levels in the brain, and its deficient expression and/or activity may be a risk factor in sporadic Alzheimer's disease (AD). Although IDE sub-cellular localization has been well studied, the compartments relevant to Abeta degradation remain to be determined.
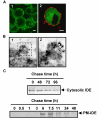
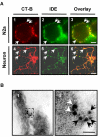
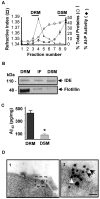
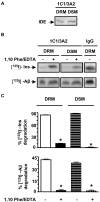
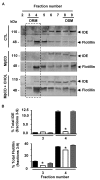
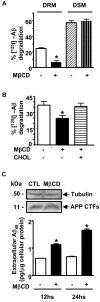
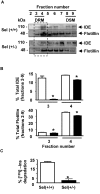
Results: Our results of live immunofluorescence, immuno gold electron-microscopy and gradient fractionation concurred to the demonstration that endogenous IDE from brain tissues and cell cultures is, in addition to its other localizations, a detergent-resistant membrane (DRM)-associated metallopeptidase. Our pulse chase experiments were in accordance with the existence of two pools of IDE: the cytosolic one with a longer half-life and the membrane-IDE with a faster turn-over. DRMs-associated IDE co-localized with Abeta and its distribution (DRMs vs. non-DRMs) and activity was sensitive to manipulation of lipid composition in vitro and in vivo. When IDE was mis-located from DRMs by treating cells with methyl-beta-cyclodextrin (MbetaCD), endogenous Abeta accumulated in the extracellular space and exogenous Abeta proteolysis was impaired. We detected a reduced amount of IDE in DRMs of membranes isolated from mice brain with endogenous reduced levels of cholesterol (Chol) due to targeted deletion of one seladin-1 allele. We confirmed that a moderate shift of IDE from DRMs induced a substantial decrement on IDE-mediated insulin and Abeta degradation in vitro.
Conclusion: Our results support the notion that optimal substrate degradation by IDE may require its association with organized-DRMs. Alternatively, DRMs but not other plasma membrane regions, may act as platforms where Abeta accumulates, due to its hydrophobic properties, reaching local concentration close to its Km for IDE facilitating its clearance. Structural integrity of DRMs may also be required to tightly retain insulin receptor and IDE for insulin proteolysis. The concept that mis-location of Abeta degrading proteases away from DRMs may impair the physiological turn-over of Abeta in vivo deserves further investigation in light of therapeutic strategies based on enhancing Abeta proteolysis in which DRM protease-targeting may need to be taken into account.
Figures

References
-
- Kurochkin IV. Insulin-degrading enzyme: embarking on amyloid destruction. Trends Biochem Sci. 2001;26:421–425. - PubMed
-
- Morelli L, Bulloj A, Leal MC, Castaño EM. Amyloid β degradation: a challenging task for brain peptidases. Subcell Biochem. 2005;38:129–145. - PubMed
-
- Duckworth WC. Insulin degradation: mechanisms, products, and significance. Endocr Rev. 1988;9:319–345. - PubMed
-
- Duckworth WC, Bennett RG, Hamel FG. Insulin degradation: progress and potential. Endocr Rev. 1998;19:608–624. - PubMed
-
- Kuo WL, Gehm BD, Rosner MR, Li W, Keller G. Inducible expression and cellular localization of insulin-degrading enzyme in a stably transfected cell line. J Biol Chem. 1994;269:22599–22606. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

