TGFBI deficiency predisposes mice to spontaneous tumor development
- PMID: 19117985
- PMCID: PMC2664305
- DOI: 10.1158/0008-5472.CAN-08-1648
TGFBI deficiency predisposes mice to spontaneous tumor development
Abstract
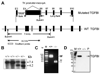
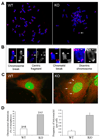
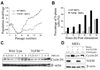
Loss of TGFBI, a secreted protein induced by transforming growth factor-beta, has been implicated in cell proliferation, tumor progression, and angiogenesis by in vitro studies. However, in vivo antitumor functions of TGFBI as well as the underlying molecular mechanism are not well understood. To these aims, we have generated a mouse model with disruption of TGFBI genomic locus. Mice lacking TGFBI show a retarded growth and are prone to spontaneous tumors and 7,12-dimethylbenz(a)anthracene-induced skin tumors. In relation to wild-type (WT) mouse embryonic fibroblasts (MEF), TGFBI(-/-) MEFs display increased frequencies of chromosomal aberration and micronuclei formation and exhibit an enhanced proliferation and early S-phase entry. Cyclin D1 is up-regulated in TGFBI(-/-) MEFs, which correlates with aberrant activation of transcription factor cyclic AMP-responsive element binding protein (CREB) identified by chromatin immunoprecipitation and luciferase reporter assays. TGFBI reconstitution in TGFBI(-/-) cells by either retroviral infection with WT TGFBI gene or supplement with recombinant mouse TGFBI protein in the culture medium leads to the suppression of CREB activation and cyclin D1 expression, and further inhibition of cell proliferation. Cyclin D1 up-regulation was also identified in most of the tumors arising from TGFBI(-/-) mice. Our studies provide the first evidence that TGFBI functions as a tumor suppressor in vivo.
Figures



References
-
- Tlsty TD. Cell-adhesion-dependent influences on genomic instability and carcinogenesis. Curr Opin Cell Biol. 1998;10:647–653. - PubMed
-
- Giancotti FG, Ruoslahti E. Integrin signaling. Science. 1999;285:1028–1032. - PubMed
-
- Danen EH. Integrins: regulators of tissue function and cancer progression. Curr Pharm Des. 2005;11:881–891. - PubMed
-
- Ramsay AG, Marshall JF, Hart IR. Integrin trafficking and its role in cancer metastasis. Cancer Metastasis Rev. 2007;26:567–578. - PubMed
-
- Skonier J, Neubauer M, Madisen L, Bennett K, Plowman GD, Purchio AF. cDNA cloning and sequence analysis of beta ig-h3, a novel gene induced in a human adenocarcinoma cell line after treatment with transforming growth factor-beta. DNA Cell Biol. 1992;11:511–522. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

