Insights into MHC class I peptide loading from the structure of the tapasin-ERp57 thiol oxidoreductase heterodimer
- PMID: 19119025
- PMCID: PMC2650231
- DOI: 10.1016/j.immuni.2008.10.018
Insights into MHC class I peptide loading from the structure of the tapasin-ERp57 thiol oxidoreductase heterodimer
Abstract
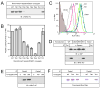
Tapasin is a glycoprotein critical for loading major histocompatibility complex (MHC) class I molecules with high-affinity peptides. It functions within the multimeric peptide-loading complex (PLC) as a disulfide-linked, stable heterodimer with the thiol oxidoreductase ERp57, and this covalent interaction is required to support optimal PLC activity. Here, we present the 2.6 A resolution structure of the tapasin-ERp57 core of the PLC. The structure revealed that tapasin interacts with both ERp57 catalytic domains, accounting for the stability of the heterodimer, and provided an example of a protein disulfide isomerase family member interacting with substrate. Mutational analysis identified a conserved surface on tapasin that interacted with MHC class I molecules and was critical for peptide loading and editing functions of the tapasin-ERp57 heterodimer. By combining the tapasin-ERp57 structure with those of other defined PLC components, we present a molecular model that illuminates the processes involved in MHC class I peptide loading.
Figures




Comment in
-
More images that yet fresh images Beget.Immunity. 2009 Jan 16;30(1):1-2. doi: 10.1016/j.immuni.2008.12.011. Immunity. 2009. PMID: 19144309
Similar articles
-
Selective loading of high-affinity peptides onto major histocompatibility complex class I molecules by the tapasin-ERp57 heterodimer.Nat Immunol. 2007 Aug;8(8):873-81. doi: 10.1038/ni1485. Epub 2007 Jul 1. Nat Immunol. 2007. PMID: 17603487
-
Tapasin and ERp57 form a stable disulfide-linked dimer within the MHC class I peptide-loading complex.EMBO J. 2005 Oct 19;24(20):3613-23. doi: 10.1038/sj.emboj.7600814. Epub 2005 Sep 29. EMBO J. 2005. PMID: 16193070 Free PMC article.
-
Major histocompatibility complex class I-ERp57-tapasin interactions within the peptide-loading complex.J Biol Chem. 2007 Jun 15;282(24):17587-93. doi: 10.1074/jbc.M702212200. Epub 2007 Apr 24. J Biol Chem. 2007. PMID: 17459881
-
Interaction of ERp57 and tapasin in the generation of MHC class I-peptide complexes.Curr Opin Immunol. 2007 Feb;19(1):99-105. doi: 10.1016/j.coi.2006.11.013. Epub 2006 Dec 5. Curr Opin Immunol. 2007. PMID: 17150345 Review.
-
The quality control of MHC class I peptide loading.Curr Opin Cell Biol. 2008 Dec;20(6):624-31. doi: 10.1016/j.ceb.2008.09.005. Epub 2008 Oct 29. Curr Opin Cell Biol. 2008. PMID: 18926908 Free PMC article. Review.
Cited by
-
Identification of rare protein disulfide isomerase gene variants in amyotrophic lateral sclerosis patients.Gene. 2015 Jul 25;566(2):158-65. doi: 10.1016/j.gene.2015.04.035. Epub 2015 Apr 22. Gene. 2015. PMID: 25913742 Free PMC article.
-
Roles of Calreticulin in Protein Folding, Immunity, Calcium Signaling and Cell Transformation.Prog Mol Subcell Biol. 2021;59:145-162. doi: 10.1007/978-3-030-67696-4_7. Prog Mol Subcell Biol. 2021. PMID: 34050865 Free PMC article.
-
Crystal structure of the HLA-DM-HLA-DR1 complex defines mechanisms for rapid peptide selection.Cell. 2012 Dec 21;151(7):1557-68. doi: 10.1016/j.cell.2012.11.025. Cell. 2012. PMID: 23260142 Free PMC article.
-
Protein disulfide isomerases contribute differentially to the endoplasmic reticulum-associated degradation of apolipoprotein B and other substrates.Mol Biol Cell. 2012 Feb;23(4):520-32. doi: 10.1091/mbc.E11-08-0704. Epub 2011 Dec 21. Mol Biol Cell. 2012. PMID: 22190736 Free PMC article.
-
Recognition Dynamics of Cancer Mutations on the ERp57-Tapasin Interface.Cancers (Basel). 2020 Mar 20;12(3):737. doi: 10.3390/cancers12030737. Cancers (Basel). 2020. PMID: 32244998 Free PMC article.
References
-
- Bangia N, Lehner PJ, Hughes EA, Surman M, Cresswell P. The N-terminal region of tapasin is required to stabilize the MHC class I loading complex. Eur J Immunol. 1998;6:1858–1870. - PubMed
-
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. - PubMed
-
- Busch R, Rinderknecht CH, Roh S, Lee AW, Harding JJ, Burster T, Hornell TM, Mellins ED. Achieving stability through editing and chaperoning: regulation of MHC class II peptide binding and expression. Immunol Rev. 2005;207:242–260. - PubMed
-
- Carreno BM, Solheim JC, Harris M, Stroynowski I, Connolly JM, Hansen TH. TAP associates with a unique class I conformation, whereas calnexin associates with multiple class I forms in mouse and man. J Immunol. 1995;155:4726–4733. - PubMed
-
- Carson M. (Academic Press) in Methods in Enzymology. 1997;277:493–505. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

