Non-coding RNA prediction and verification in Saccharomyces cerevisiae
- PMID: 19119416
- PMCID: PMC2603021
- DOI: 10.1371/journal.pgen.1000321
Non-coding RNA prediction and verification in Saccharomyces cerevisiae
Abstract
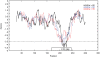
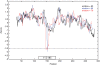
Non-coding RNA (ncRNA) play an important and varied role in cellular function. A significant amount of research has been devoted to computational prediction of these genes from genomic sequence, but the ability to do so has remained elusive due to a lack of apparent genomic features. In this work, thermodynamic stability of ncRNA structural elements, as summarized in a Z-score, is used to predict ncRNA in the yeast Saccharomyces cerevisiae. This analysis was coupled with comparative genomics to search for ncRNA genes on chromosome six of S. cerevisiae and S. bayanus. Sets of positive and negative control genes were evaluated to determine the efficacy of thermodynamic stability for discriminating ncRNA from background sequence. The effect of window sizes and step sizes on the sensitivity of ncRNA identification was also explored. Non-coding RNA gene candidates, common to both S. cerevisiae and S. bayanus, were verified using northern blot analysis, rapid amplification of cDNA ends (RACE), and publicly available cDNA library data. Four ncRNA transcripts are well supported by experimental data (RUF10, RUF11, RUF12, RUF13), while one additional putative ncRNA transcript is well supported but the data are not entirely conclusive. Six candidates appear to be structural elements in 5' or 3' untranslated regions of annotated protein-coding genes. This work shows that thermodynamic stability, coupled with comparative genomics, can be used to predict ncRNA with significant structural elements.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Comparative analysis of structured RNAs in S. cerevisiae indicates a multitude of different functions.BMC Biol. 2007 Jun 18;5:25. doi: 10.1186/1741-7007-5-25. BMC Biol. 2007. PMID: 17577407 Free PMC article.
-
Computational identification of non-coding RNAs in Saccharomyces cerevisiae by comparative genomics.Nucleic Acids Res. 2003 Jul 15;31(14):4119-28. doi: 10.1093/nar/gkg438. Nucleic Acids Res. 2003. PMID: 12853629 Free PMC article.
-
Degradation of Non-coding RNAs Promotes Recycling of Termination Factors at Sites of Transcription.Cell Rep. 2020 Jul 21;32(3):107942. doi: 10.1016/j.celrep.2020.107942. Cell Rep. 2020. PMID: 32698007
-
Identification and annotation of noncoding RNAs in Saccharomycotina.C R Biol. 2011 Aug-Sep;334(8-9):671-8. doi: 10.1016/j.crvi.2011.05.016. Epub 2011 Jul 6. C R Biol. 2011. PMID: 21819949 Review.
-
Non-coding transcription by RNA polymerase II in yeast: Hasard or nécessité?Biochimie. 2015 Oct;117:28-36. doi: 10.1016/j.biochi.2015.04.020. Epub 2015 May 6. Biochimie. 2015. PMID: 25956976 Review.
Cited by
-
Regulatory RNAs in brain function and disorders.Brain Res. 2010 Jun 18;1338:36-47. doi: 10.1016/j.brainres.2010.03.042. Epub 2010 Mar 20. Brain Res. 2010. PMID: 20307503 Free PMC article. Review.
-
The conserved chimeric transcript UPGRADE2 is associated with unreduced pollen formation and is exclusively found in apomictic Boechera species.Plant Physiol. 2013 Dec;163(4):1640-59. doi: 10.1104/pp.113.222448. Epub 2013 Oct 15. Plant Physiol. 2013. PMID: 24130193 Free PMC article.
-
RNAStructuromeDB: A genome-wide database for RNA structural inference.Sci Rep. 2017 Dec 8;7(1):17269. doi: 10.1038/s41598-017-17510-y. Sci Rep. 2017. PMID: 29222504 Free PMC article.
-
Large-scale profiling of noncoding RNA function in yeast.PLoS Genet. 2018 Mar 12;14(3):e1007253. doi: 10.1371/journal.pgen.1007253. eCollection 2018 Mar. PLoS Genet. 2018. PMID: 29529031 Free PMC article.
-
Functional annotations for the Saccharomyces cerevisiae genome: the knowns and the known unknowns.Trends Microbiol. 2009 Jul;17(7):286-94. doi: 10.1016/j.tim.2009.04.005. Epub 2009 Jul 2. Trends Microbiol. 2009. PMID: 19577472 Free PMC article. Review.
References
-
- Eddy SR. Non-coding RNA genes and the modern RNA world. Nat Rev Genet. 2001;2:919–929. - PubMed
-
- Storz G. An expanding universe of noncoding RNAs. Science. 2002;296:1260–1263. - PubMed
-
- Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15 Spec No 1:R17–R29. - PubMed
-
- Costa FF. Non-coding RNAs: lost in translation? Gene. 2007;386:1–10. - PubMed
-
- Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

