Positive selection results in frequent reversible amino acid replacements in the G protein gene of human respiratory syncytial virus
- PMID: 19119418
- PMCID: PMC2603285
- DOI: 10.1371/journal.ppat.1000254
Positive selection results in frequent reversible amino acid replacements in the G protein gene of human respiratory syncytial virus
Abstract
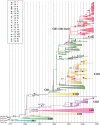
Human respiratory syncytial virus (HRSV) is the major cause of lower respiratory tract infections in children under 5 years of age and the elderly, causing annual disease outbreaks during the fall and winter. Multiple lineages of the HRSVA and HRSVB serotypes co-circulate within a single outbreak and display a strongly temporal pattern of genetic variation, with a replacement of dominant genotypes occurring during consecutive years. In the present study we utilized phylogenetic methods to detect and map sites subject to adaptive evolution in the G protein of HRSVA and HRSVB. A total of 29 and 23 amino acid sites were found to be putatively positively selected in HRSVA and HRSVB, respectively. Several of these sites defined genotypes and lineages within genotypes in both groups, and correlated well with epitopes previously described in group A. Remarkably, 18 of these positively selected tended to revert in time to a previous codon state, producing a "flip-flop" phylogenetic pattern. Such frequent evolutionary reversals in HRSV are indicative of a combination of frequent positive selection, reflecting the changing immune status of the human population, and a limited repertoire of functionally viable amino acids at specific amino acid sites.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Collins PL, Chanock RM, Murphy BR. Respiratory syncytial virus, In: Kinipe DM, Howley PM, Griffin DE, Lamb RA, editors. Philadelphia, , Pa: Lippicncott Williams and Eilkins; 2001. pp. 1443–1485.
-
- Falsey AR, Hennessey PA, Formica MA, Cox C, Walsh EE. Respiratory syncytial virus infection in elderly and high-risk adults. New England Journal of Medicine. 2005;52(17):1749–59. - PubMed
-
- Ison MG, Hayden FG. Viral infections in immunocompromised patients: what's new with respiratory viruses? Curr Opin Infect Dis. 2002;15:355–367. - PubMed
-
- Anderson LJ, Hierholzer JC, Tsou C, Hendry RM, Fernie BF, et al. Antigenic characterization of respiratory syncytial virus strains with monoclonal antibodies. J Infect Dis. 1985;151:626–633. - PubMed
-
- Mufson MA, Örvell C, Rafnar B, Norrby E. Two distinct subtypes of human respiratory syncytial virus. J Gen Virol. 1985;66:2111–2124, 1985. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

