A novel role for Mc1r in the parallel evolution of depigmentation in independent populations of the cavefish Astyanax mexicanus
- PMID: 19119422
- PMCID: PMC2603666
- DOI: 10.1371/journal.pgen.1000326
A novel role for Mc1r in the parallel evolution of depigmentation in independent populations of the cavefish Astyanax mexicanus
Abstract
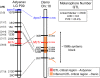
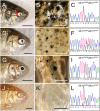
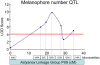
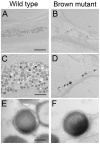
The evolution of degenerate characteristics remains a poorly understood phenomenon. Only recently has the identification of mutations underlying regressive phenotypes become accessible through the use of genetic analyses. Focusing on the Mexican cave tetra Astyanax mexicanus, we describe, here, an analysis of the brown mutation, which was first described in the literature nearly 40 years ago. This phenotype causes reduced melanin content, decreased melanophore number, and brownish eyes in convergent cave forms of A. mexicanus. Crosses demonstrate non-complementation of the brown phenotype in F(2) individuals derived from two independent cave populations: Pachón and the linked Yerbaniz and Japonés caves, indicating the same locus is responsible for reduced pigmentation in these fish. While the brown mutant phenotype arose prior to the fixation of albinism in Pachón cave individuals, it is unclear whether the brown mutation arose before or after the fixation of albinism in the linked Yerbaniz/Japonés caves. Using a QTL approach combined with sequence and functional analyses, we have discovered that two distinct genetic alterations in the coding sequence of the gene Mc1r cause reduced pigmentation associated with the brown mutant phenotype in these caves. Our analysis identifies a novel role for Mc1r in the evolution of degenerative phenotypes in blind Mexican cavefish. Further, the brown phenotype has arisen independently in geographically separate caves, mediated through different mutations of the same gene. This example of parallelism indicates that certain genes are frequent targets of mutation in the repeated evolution of regressive phenotypes in cave-adapted species.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



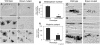

References
-
- Protas ME, Hersey C, Kochanek D, Zhou Y, Wilkens H, et al. Genetic analysis of cavefish reveals molecular convergence in the evolution of albinism. Nat Genet. 2006;38:107–111. - PubMed
-
- Protas M, Tabansky I, Conrad M, Gross JB, Vidal O, et al. Multi-trait evolution in a cave fish, Astyanax mexicanus. Evol Dev. 2008;10:196–209. - PubMed
-
- Bensouilah M, Denizot J-P. Taste buds and neuromasts of Astyanax jordani: Distribution and immunochemical demonstration of co-localized substance P and enkephalins. Eur J Neurosci. 1991;3:407–414. - PubMed
-
- Wilkens H. Evolution and genetics of epigean and cave Astyanax fasciatus (Characidae, Pisces): Support for the neutral mutation theory. In: Hecht MK, Wallace B, editors. Evolutionary biology. New York: Plenum Publishing Corporation; 1988. pp. 271–367.
-
- Jeffery WR. Cavefish as a model system in evolutionary developmental biology. Dev Biol. 2001;231:1–12. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

