Adaptive evolution in zinc finger transcription factors
- PMID: 19119423
- PMCID: PMC2604467
- DOI: 10.1371/journal.pgen.1000325
Adaptive evolution in zinc finger transcription factors
Abstract
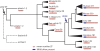
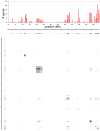
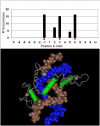
The majority of human genes are conserved among mammals, but some gene families have undergone extensive expansion in particular lineages. Here, we present an evolutionary analysis of one such gene family, the poly-zinc-finger (poly-ZF) genes. The human genome encodes approximately 700 members of the poly-ZF family of putative transcriptional repressors, many of which have associated KRAB, SCAN, or BTB domains. Analysis of the gene family across the tree of life indicates that the gene family arose from a small ancestral group of eukaryotic zinc-finger transcription factors through many repeated gene duplications accompanied by functional divergence. The ancestral gene family has probably expanded independently in several lineages, including mammals and some fishes. Investigation of adaptive evolution among recent paralogs using d(N)/d(S) analysis indicates that a major component of the selective pressure acting on these genes has been positive selection to change their DNA-binding specificity. These results suggest that the poly-ZF genes are a major source of new transcriptional repression activity in humans and other primates.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Carroll SB, Grenier JK, Weatherbee SD. From DNA to Diversity, 2nd edn. Malden, MA: Blackwell Publishing; 2005.
-
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. - PubMed
-
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, et al. The sequence of the human genome. Science. 2001;291(5507):1304–1351. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

