Autophagy and pattern recognition receptors in innate immunity
- PMID: 19120485
- PMCID: PMC2788953
- DOI: 10.1111/j.1600-065X.2008.00725.x
Autophagy and pattern recognition receptors in innate immunity
Abstract
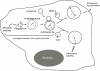
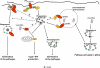
Autophagy is a physiologically and immunologically controlled intracellular homeostatic pathway that sequesters and degrades cytoplasmic targets including macromolecular aggregates, cellular organelles such as mitochondria, and whole microbes or their products. Recent advances show that autophagy plays a role in innate immunity in several ways: (i) direct elimination of intracellular microbes by digestion in autolysosomes, (ii) delivery of cytosolic microbial products to pattern recognition receptors (PRRs) in a process referred to as topological inversion, and (iii) as an anti-microbial effector of Toll-like receptors and other PRR signaling. Autophagy eliminates pathogens in vitro and in vivo but, when aberrant due to mutations, contributes to human inflammatory disorders such as Crohn's disease. In this review, we examine these relationships and propose that autophagy is one of the most ancient innate immune defenses that has possibly evolved at the time of alpha-protobacteria-pre-eukaryote relationships, leading up to modern eukaryotic cell-mitochondrial symbiosis, and that during the metazoan evolution, additional layers of immunological regulation have been superimposed and integrated with this primordial innate immunity mechanism.
Figures



Similar articles
-
Multiple regulatory and effector roles of autophagy in immunity.Curr Opin Immunol. 2009 Feb;21(1):53-62. doi: 10.1016/j.coi.2009.02.002. Epub 2009 Mar 5. Curr Opin Immunol. 2009. PMID: 19269148 Free PMC article.
-
Autophagy in immunity against mycobacterium tuberculosis: a model system to dissect immunological roles of autophagy.Curr Top Microbiol Immunol. 2009;335:169-88. doi: 10.1007/978-3-642-00302-8_8. Curr Top Microbiol Immunol. 2009. PMID: 19802565 Free PMC article. Review.
-
Intracellular recognition of pathogens and autophagy as an innate immune host defence.J Biochem. 2011 Aug;150(2):143-9. doi: 10.1093/jb/mvr083. Epub 2011 Jul 5. J Biochem. 2011. PMID: 21729928 Free PMC article. Review.
-
Links between autophagy, innate immunity, inflammation and Crohn's disease.Dig Dis. 2009;27(3):246-51. doi: 10.1159/000228557. Epub 2009 Sep 24. Dig Dis. 2009. PMID: 19786748 Free PMC article. Review.
-
Toll-like receptors in control of immunological autophagy.Cell Death Differ. 2009 Jul;16(7):976-83. doi: 10.1038/cdd.2009.40. Epub 2009 May 15. Cell Death Differ. 2009. PMID: 19444282 Free PMC article. Review.
Cited by
-
Autophagy: New Questions from Recent Answers.ISRN Mol Biol. 2012 Dec 30;2012:738718. doi: 10.5402/2012/738718. eCollection 2012. ISRN Mol Biol. 2012. PMID: 27335669 Free PMC article. Review.
-
MicroRNA-155 promotes autophagy to eliminate intracellular mycobacteria by targeting Rheb.PLoS Pathog. 2013;9(10):e1003697. doi: 10.1371/journal.ppat.1003697. Epub 2013 Oct 10. PLoS Pathog. 2013. PMID: 24130493 Free PMC article.
-
Pseudomonas aeruginosa Triggers Macrophage Autophagy To Escape Intracellular Killing by Activation of the NLRP3 Inflammasome.Infect Immun. 2015 Oct 14;84(1):56-66. doi: 10.1128/IAI.00945-15. Print 2016 Jan. Infect Immun. 2015. PMID: 26467446 Free PMC article.
-
Human papillomavirus infection is inhibited by host autophagy in primary human keratinocytes.Virology. 2013 Mar 1;437(1):12-9. doi: 10.1016/j.virol.2012.12.004. Epub 2013 Jan 4. Virology. 2013. PMID: 23290079 Free PMC article.
-
Lipopolysaccharide stimulates p62-dependent autophagy-like aggregate clearance in hepatocytes.Biomed Res Int. 2014;2014:267350. doi: 10.1155/2014/267350. Epub 2014 Feb 10. Biomed Res Int. 2014. PMID: 24683544 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

