Cytokinesis is blocked in mammalian cells transfected with Chlamydia trachomatis gene CT223
- PMID: 19123944
- PMCID: PMC2657910
- DOI: 10.1186/1471-2180-9-2
Cytokinesis is blocked in mammalian cells transfected with Chlamydia trachomatis gene CT223
Abstract
Background: The chlamydiae alter many aspects of host cell biology, including the division process, but the molecular biology of these alterations remains poorly characterized. Chlamydial inclusion membrane proteins (Incs) are likely candidates for direct interactions with host cell cytosolic proteins, as they are secreted to the inclusion membrane and exposed to the cytosol. The inc gene CT223 is one of a sequential set of orfs that encode or are predicted to encode Inc proteins. CT223p is localized to the inclusion membrane in all tested C. trachomatis serovars.
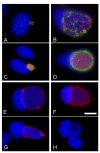
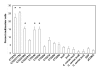
Results: A plasmid transfection approach was used to examine the function of the product of CT223 and other Inc proteins within uninfected mammalian cells. Fluorescence microscopy was used to demonstrate that CT223, and, to a lesser extent, adjacent inc genes, are capable of blocking host cell cytokinesis and facilitating centromere supranumeracy defects seen by others in chlamydiae-infected cells. Both phenotypes were associated with transfection of plasmids encoding the carboxy-terminal tail of CT223p, a region of the protein that is likely exposed to the cytosol in infected cells.
Conclusion: These studies suggest that certain Inc proteins block cytokinesis in C. trachomatis-infected cells. These results are consistent with the work of others showing chlamydial inhibition of host cell cytokinesis.
Figures




References
-
- Valdivia RH. Chlamydia effector proteins and new insights into chlamydial cellular microbiology. Curr Opin Microbiol. 2008;11(1):53–59. - PubMed
-
- Fields KA, Hackstadt T. The chlamydial inclusion: escape from the endocytic pathway. Annu Rev Cell Dev Biol. 2002;18:221–245. - PubMed
-
- Mabey D. Trachoma: recent developments. Adv Exp Med Biol. 2008;609:98–107. - PubMed
-
- Stamm WE. Chlamydia trachomatis infections: progress and problems. J Infect Dis. 1999;179(Suppl 2):S380–383. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

