TRF1 controls telomere length and mitotic fidelity in epithelial homeostasis
- PMID: 19124610
- PMCID: PMC2648247
- DOI: 10.1128/MCB.01339-08
TRF1 controls telomere length and mitotic fidelity in epithelial homeostasis
Abstract
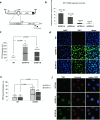
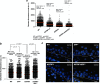
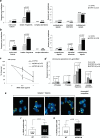
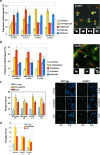
TRF1 is a component of the shelterin complex at mammalian telomeres; however, a role for TRF1 in telomere biology in the context of the organism is unclear. In this study, we generated mice with transgenic TRF1 expression targeted to epithelial tissues (K5TRF1 mice). K5TRF1 mice have shorter telomeres in the epidermis than wild-type controls do, and these are rescued in the absence of the XPF nuclease, indicating that TRF1 acts as a negative regulator of telomere length by controlling XPF activity at telomeres, similar to what was previously described for TRF2-overexpressing mice (K5TRF2 mice). K5TRF1 cells also show increased end-to-end chromosomal fusions, multitelomeric signals, and increased telomere recombination, indicating an impact of TRF1 on telomere integrity, again similar to the case in K5TRF2 cells. Intriguingly, K5TRF1 cells, but not K5TRF2 cells, show increased mitotic spindle aberrations. TRF1 colocalizes with the spindle assembly checkpoint proteins BubR1 and Mad2 at mouse telomeres, indicating a link between telomeres and the mitotic spindle. Together, these results demonstrate that TRF1, like TRF2, negatively regulates telomere length in vivo by controlling the action of the XPF nuclease at telomeres; in addition, TRF1 has a unique role in the mitotic spindle checkpoint.
Figures








Similar articles
-
TopoIIα prevents telomere fragility and formation of ultra thin DNA bridges during mitosis through TRF1-dependent binding to telomeres.Cell Cycle. 2014;13(9):1463-81. doi: 10.4161/cc.28419. Epub 2014 Mar 10. Cell Cycle. 2014. PMID: 24626180 Free PMC article.
-
Increased telomere fragility and fusions resulting from TRF1 deficiency lead to degenerative pathologies and increased cancer in mice.Genes Dev. 2009 Sep 1;23(17):2060-75. doi: 10.1101/gad.543509. Epub 2009 Aug 13. Genes Dev. 2009. PMID: 19679647 Free PMC article.
-
The telomere repeat binding protein Trf1 interacts with the spindle checkpoint protein Mad1 and Nek2 mitotic kinase.Cell Cycle. 2005 Jan;4(1):121-4. doi: 10.4161/cc.4.1.1351. Epub 2005 Jan 5. Cell Cycle. 2005. PMID: 15611654
-
Role of Pin2/TRF1 in telomere maintenance and cell cycle control.J Cell Biochem. 2003 May 1;89(1):19-37. doi: 10.1002/jcb.10496. J Cell Biochem. 2003. PMID: 12682905 Review.
-
Post-translational modifications of TRF1 and TRF2 and their roles in telomere maintenance.Mech Ageing Dev. 2012 Jun;133(6):421-34. doi: 10.1016/j.mad.2012.05.002. Epub 2012 May 23. Mech Ageing Dev. 2012. PMID: 22634377 Review.
Cited by
-
Generation of mice with longer and better preserved telomeres in the absence of genetic manipulations.Nat Commun. 2016 Jun 2;7:11739. doi: 10.1038/ncomms11739. Nat Commun. 2016. PMID: 27252083 Free PMC article.
-
Exercise alters mRNA expression of telomere-repeat binding factor 1 in skeletal muscle via p38 MAPK.J Appl Physiol (1985). 2012 Dec 1;113(11):1737-46. doi: 10.1152/japplphysiol.00200.2012. Epub 2012 Oct 4. J Appl Physiol (1985). 2012. PMID: 23042912 Free PMC article.
-
The 68-kDa telomeric repeat binding factor 1 (TRF1)-associated protein (TAP68) interacts with and recruits TRF1 to the spindle pole during mitosis.J Biol Chem. 2014 May 16;289(20):14145-56. doi: 10.1074/jbc.M113.526244. Epub 2014 Apr 1. J Biol Chem. 2014. PMID: 24692559 Free PMC article.
-
Telomerase Inhibition by MST-312 Sensitizes Breast Cancer Cells to the Anti-cancer Properties of Plumbagin.Genome Integr. 2023 Sep 12;14:e20230002. doi: 10.14293/genint.14.1.003. eCollection 2023. Genome Integr. 2023. PMID: 38765717 Free PMC article.
-
E-type cyclins modulate telomere integrity in mammalian male meiosis.Chromosoma. 2016 Jun;125(2):253-64. doi: 10.1007/s00412-015-0564-3. Epub 2015 Dec 28. Chromosoma. 2016. PMID: 26712234 Free PMC article.
References
-
- Ancelin, K., M. Brunori, S. Bauwens, C. E. Koering, C. Brun, M. Ricoul, J. P. Pommier, L. Sabatier, and E. Gilson. 2002. Targeting assay to study the cis functions of human telomeric proteins: evidence for inhibition of telomerase by TRF1 and for activation of telomere degradation by TRF2. Mol. Cell. Biol. 223474-3487. - PMC - PubMed
-
- Azzalin, C. M., P. Reichenbach, L. Khoriauli, E. Giulotto, and J. Lingner. 2007. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318798-801. - PubMed
-
- Bechter, O. E., Y. Zou, W. Walker, W. E. Wright, and J. W. Shay. 2004. Telomeric recombination in mismatch repair deficient human colon cancer cells after telomerase inhibition. Cancer Res. 643444-3451. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
