Alternative splicing: regulation without regulators
- PMID: 19125169
- PMCID: PMC2732116
- DOI: 10.1038/nsmb0109-13
Alternative splicing: regulation without regulators
Abstract
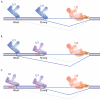
Alternative splicing is typically thought to be controlled by RNA binding proteins that modulate the activity of the spliceosome. A new study not only demonstrates that alternative splicing can be regulated without the involvement of auxiliary splicing factors, but also provides mechanistic insight into how this can occur.
Figures

References
-
- Wang, et al. Nature. 2008 doi:10.1038/nature07509.
-
- Yu, et al. Cell. in press.
-
- Lin S, Fu XD. In: Alternative Splicing in the Postgenomic Era. Blencowe BJ, Graveley BR, editors. Landes Bioscience and Springer Science+Business Media, LLC; New York: 2007. pp. 107–122.
-
- Martinez-Contreras R, et al. In: Alternative Splicing in the Postgenomic Era. Blencowe BJ, Graveley BR, editors. Landes Bioscience and Springer Science+Business Media, LLC; New York: 2007. pp. 123–147.
-
- Wang, et al. Cell. 2004;119:831–845. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

