RNA interference screening demystified
- PMID: 19126568
- PMCID: PMC2652024
- DOI: 10.1136/jcp.2008.058735
RNA interference screening demystified
Abstract
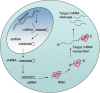
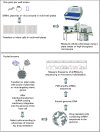
Genetic screens, where the effects of modifying gene function on cell behaviour are assessed in a systematic fashion, have for some time provided useful information to those interested in disease pathogenesis and treatment. Genetic screens exploiting the phenomenon of RNA interference (RNAi) are now becoming commonplace. This article explains the different RNAi screen formats and describes some of the applications of RNAi screening that may be pertinent to the research pathologist.
Conflict of interest statement
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
