Possible diversifying selection in the imprinted gene, MEDEA, in Arabidopsis
- PMID: 19126870
- PMCID: PMC2727397
- DOI: 10.1093/molbev/msp001
Possible diversifying selection in the imprinted gene, MEDEA, in Arabidopsis
Abstract
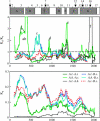
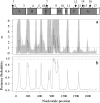
Coevolutionary conflict among imprinted genes that influence traits such as offspring growth may arise when maternal and paternal genomes have different evolutionary optima. This conflict is expected in outcrossing taxa with multiple paternity, but not self-fertilizing taxa. MEDEA (MEA) is an imprinted plant gene that influences seed growth. Disagreement exists regarding the type of selection acting on this gene. We present new data and analyses of sequence diversity of MEA in self-fertilizing and outcrossing Arabidopsis and its relatives, to help clarify the form of selection acting on this gene. Codon-based branch analysis among taxa (PAML) suggests that selection on the coding region is changing over time, and nonsynonymous substitution is elevated in at least one outcrossing branch. Codon-based analysis of diversity within outcrossing Arabidopsis lyrata ssp. petraea (OmegaMap) suggests that diversifying selection is acting on a portion of the gene, to cause elevated nonsynonymous polymorphism. Providing further support for balancing selection in A. lyrata, Hudson, Kreitman and Aguadé analysis indicates that diversity/divergence at silent sites in the MEA promoter and genic region is elevated relative to reference genes, and there are deviations from the neutral frequency spectrum. This combination of positive selection as well as balancing and diversifying selection in outcrossing lineages is consistent with other genes influence by evolutionary conflict, such as disease resistance genes. Consistent with predictions that conflict would be eliminated in self-fertilizing taxa, we found no evidence of positive, balancing, or diversifying selection in A. thaliana promoter or genic region.
Figures


Similar articles
-
Paternally Expressed Imprinted Genes under Positive Darwinian Selection in Arabidopsis thaliana.Mol Biol Evol. 2019 Jun 1;36(6):1239-1253. doi: 10.1093/molbev/msz063. Mol Biol Evol. 2019. PMID: 30913563 Free PMC article.
-
High diversity due to balancing selection in the promoter region of the Medea gene in Arabidopsis lyrata.Curr Biol. 2007 Nov 6;17(21):1885-9. doi: 10.1016/j.cub.2007.09.051. Epub 2007 Oct 18. Curr Biol. 2007. PMID: 17949979
-
Positive darwinian selection at the imprinted MEDEA locus in plants.Nature. 2007 Jul 19;448(7151):349-52. doi: 10.1038/nature05984. Nature. 2007. PMID: 17637669
-
[Genomic imprinting and seed development].Yi Chuan. 2005 Jul;27(4):665-70. Yi Chuan. 2005. PMID: 16120596 Review. Chinese.
-
Genomic imprinting during seed development.Adv Genet. 2002;46:165-214. doi: 10.1016/s0065-2660(02)46007-5. Adv Genet. 2002. PMID: 11931224 Review.
Cited by
-
Paternally Expressed Imprinted Genes under Positive Darwinian Selection in Arabidopsis thaliana.Mol Biol Evol. 2019 Jun 1;36(6):1239-1253. doi: 10.1093/molbev/msz063. Mol Biol Evol. 2019. PMID: 30913563 Free PMC article.
-
Conserved noncoding sequences and de novo Mutator insertion alleles are imprinted in maize.Plant Physiol. 2023 Jan 2;191(1):299-316. doi: 10.1093/plphys/kiac459. Plant Physiol. 2023. PMID: 36173333 Free PMC article.
-
The evolution of genomic imprinting: theories, predictions and empirical tests.Heredity (Edinb). 2014 Aug;113(2):119-28. doi: 10.1038/hdy.2014.29. Epub 2014 Apr 23. Heredity (Edinb). 2014. PMID: 24755983 Free PMC article. Review.
-
Comprehensive analysis of imprinted genes in maize reveals allelic variation for imprinting and limited conservation with other species.Proc Natl Acad Sci U S A. 2013 Nov 26;110(48):19639-44. doi: 10.1073/pnas.1309182110. Epub 2013 Nov 11. Proc Natl Acad Sci U S A. 2013. PMID: 24218619 Free PMC article.
-
Progress and Promise in using Arabidopsis to Study Adaptation, Divergence, and Speciation.Arabidopsis Book. 2010;8:e0138. doi: 10.1199/tab.0138. Epub 2010 Sep 29. Arabidopsis Book. 2010. PMID: 22303263 Free PMC article.
References
-
- Abi-Rached L, Dorighi K, Norman PJ, Yawata M, Parham P. Episodes of natural selection shaped the interactions of IgA-Fc with FcαRI and bacterial decoy proteins. J Immunol. 2007;178:7943–7954. - PubMed
-
- Agrawal AF, Brodie ED, Brown J. Parent–offspring coadaptation and the dual genetic control of maternal care. Science. 2001;292:1710–1712. - PubMed
-
- Andrés JA, Maroja LS, Bogdanowicz SM, Swanson WJ, Harrison RG. Molecular evolution of seminal proteins in field crickets. Mol Biol Evol. 2006;23:1574–1584. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

