Genes involved in arsenic transformation and resistance associated with different levels of arsenic-contaminated soils
- PMID: 19128515
- PMCID: PMC2631446
- DOI: 10.1186/1471-2180-9-4
Genes involved in arsenic transformation and resistance associated with different levels of arsenic-contaminated soils
Abstract
Background: Arsenic is known as a toxic metalloid, which primarily exists in inorganic form [As(III) and As(V)] and can be transformed by microbial redox processes in the natural environment. As(III) is much more toxic and mobile than As(V), hence microbial arsenic redox transformation has a major impact on arsenic toxicity and mobility which can greatly influence the human health. Our main purpose was to investigate the distribution and diversity of microbial arsenite-resistant species in three different arsenic-contaminated soils, and further study the As(III) resistance levels and related functional genes of these species.
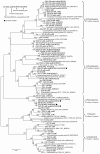
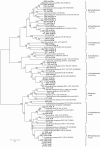
Results: A total of 58 arsenite-resistant bacteria were identified from soils with three different arsenic-contaminated levels. Highly arsenite-resistant bacteria (MIC > 20 mM) were only isolated from the highly arsenic-contaminated site and belonged to Acinetobacter, Agrobacterium, Arthrobacter, Comamonas, Rhodococcus, Stenotrophomonas and Pseudomonas. Five arsenite-oxidizing bacteria that belonged to Achromobacter, Agrobacterium and Pseudomonas were identified and displayed a higher average arsenite resistance level than the non-arsenite oxidizers. 5 aoxB genes encoding arsenite oxidase and 51 arsenite transporter genes [18 arsB, 12 ACR3(1) and 21 ACR3(2)] were successfully amplified from these strains using PCR with degenerate primers. The aoxB genes were specific for the arsenite-oxidizing bacteria. Strains containing both an arsenite oxidase gene (aoxB) and an arsenite transporter gene (ACR3 or arsB) displayed a higher average arsenite resistance level than those possessing an arsenite transporter gene only. Horizontal transfer of ACR3(2) and arsB appeared to have occurred in strains that were primarily isolated from the highly arsenic-contaminated soil.
Conclusion: Soils with long-term arsenic contamination may result in the evolution of highly diverse arsenite-resistant bacteria and such diversity was probably caused in part by horizontal gene transfer events. Bacteria capable of both arsenite oxidation and arsenite efflux mechanisms had an elevated arsenite resistance level.
Figures

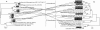
References
-
- Valls M, de Lorenzo V. Exploiting the genetic and biochemical capacities of bacteria for the remediation of heavy metal pollution. FEMS Microbiol Rev. 2002;26(4):327–338. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

