PanCGH: a genotype-calling algorithm for pangenome CGH data
- PMID: 19129208
- PMCID: PMC2639077
- DOI: 10.1093/bioinformatics/btn632
PanCGH: a genotype-calling algorithm for pangenome CGH data
Abstract
Motivation: Pangenome arrays contain DNA oligomers targeting several sequenced reference genomes from the same species. In microbiology, these can be employed to investigate the often high genetic variability within a species by comparative genome hybridization (CGH). The biological interpretation of pangenome CGH data depends on the ability to compare strains at a functional level, particularly by comparing the presence or absence of orthologous genes. Due to the high genetic variability, available genotype-calling algorithms can not be applied to pangenome CGH data.
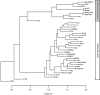
Results: We have developed the algorithm PanCGH that incorporates orthology information about genes to predict the presence or absence of orthologous genes in a query organism using CGH arrays that target the genomes of sequenced representatives of a group of microorganisms. PanCGH was tested and applied in the analysis of genetic diversity among 39 Lactococcus lactis strains from three different subspecies (lactis.cremoris, hordniae) and isolated from two different niches (dairy and plant). Clustering of these strains using the presence/absence data of gene orthologs revealed a clear separation between different subspecies and reflected the niche of the strains.
Figures
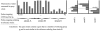
References
-
- Cleveland WS, et al. Local regression models. In: Chambers JM, Hastie TJ, editors. Chapter 8 of Statistical Models in S. Cole: Wadsworth & Brooks; 1992. pp. 312–316.
-
- Fields Development Team. Fields: Tools for Spatial Data. National Center for Atmospheric Research, Boulder, CO: 2006. [(last accessed August, 2008)]. Available at http://www.image.ucar.edu/Software/Fields/
-
- Fitch WM. Distinguishing homologous from analogous proteins. Syst. Zool. 1970;19:99–113. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

