Transcriptional control by adenovirus E1A conserved region 3 via p300/CBP
- PMID: 19129215
- PMCID: PMC2651774
- DOI: 10.1093/nar/gkn1057
Transcriptional control by adenovirus E1A conserved region 3 via p300/CBP
Abstract
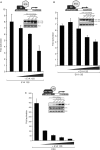
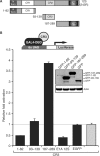
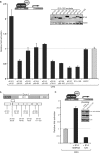
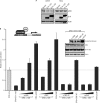
The human adenovirus type 5 (HAdV-5) E1A 13S oncoprotein is a potent regulator of gene expression and is used extensively as a model for transcriptional activation. It possesses two independent transcriptional activation domains located in the N-terminus/conserved region (CR) 1 and CR3. The protein acetyltransferase p300 was previously identified by its association with the N-terminus/CR1 portion of E1A and this association is required for oncogenic transformation by E1A. We report here that transcriptional activation by 13S E1A is inhibited by co-expression of sub-stoichiometric amounts of the smaller 12S E1A isoform, which lacks CR3. Transcriptional inhibition by E1A 12S maps to the N-terminus and correlates with the ability to bind p300/CBP, suggesting that E1A 12S is sequestering this limiting factor from 13S E1A. This is supported by the observation that the repressive effect of E1A 12S is reversed by expression of exogenous p300 or CBP, but not by a CBP mutant lacking actyltransferase activity. Furthermore, we show that transcriptional activation by 13S E1A is greatly reduced by siRNA knockdown of p300 and that CR3 binds p300 independently of the well-characterized N-terminal/CR1-binding site. Importantly, CR3 is also required to recruit p300 to the adenovirus E4 promoter during infection. These results identify a new functionally significant interaction between E1A CR3 and the p300/CBP acetyltransferases, expanding our understanding of the mechanism by which this potent transcriptional activator functions.
Figures



References
-
- Berk AJ. Recent lessons in gene expression, cell cycle control, and cell biology from adenovirus. Oncogene. 2005;24:7673–7685. - PubMed
-
- Bayley ST, Mymryk JS. Adenovirus E1A proteins and transformation (Review) Int. J. Oncology. 1994;5:425–444. - PubMed
-
- Avvakumov N, Kajon AE, Hoeben RC, Mymryk JS. Comprehensive sequence analysis of the E1A proteins of human and simian adenoviruses. Virology. 2004;329:477–492. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

