Identification of an inter-transcription factor regulatory network in human hepatoma cells by Matrix RNAi
- PMID: 19129217
- PMCID: PMC2651797
- DOI: 10.1093/nar/gkn1028
Identification of an inter-transcription factor regulatory network in human hepatoma cells by Matrix RNAi
Abstract
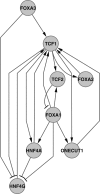
Transcriptional regulation by transcriptional regulatory factors (TRFs) of their target TRF genes is central to the control of gene expression. To study a static multi-tiered inter-TRF regulatory network in the human hepatoma cells, we have applied a Matrix RNAi approach in which siRNA knockdown and quantitative RT-PCR are used in combination on the same set of TRFs to determine their interdependencies. This approach focusing on several liver-enriched TRF families, each of which consists of structurally homologous members, revealed many significant regulatory relationships. These include the cross-talks between hepatocyte nuclear factors (HNFs) and the other TRF groups such as CCAAT/enhancer-binding proteins (CEBPs), retinoic acid receptors (RARs), retinoid receptors (RXRs) and RAR-related orphan receptors (RORs), which play key regulatory functions in human hepatocytes and liver. In addition, various multi-component regulatory motifs, which make up the complex inter-TRF regulatory network, were identified. A large part of the regulatory edges identified by the Matrix RNAi approach could be confirmed by chromatin immunoprecipitation. The resultant significant edges enabled us to depict the inter-TRF TRN forming an apparent regulatory hierarchy of (FOXA1, RXRA) --> TCF1 --> (HNF4A, ONECUT1) --> (RORC, CEBPA) as the main streamline.
Figures








References
-
- Simon I, Barnett J, Hannett N, Harbison CT, Rinaldi NJ, Volkert TL, Wyrick JJ, Zeitlinger J, Gifford DK, Jaakkola TS, et al. Serial regulation of transcriptional regulators in the yeast cell cycle. Cell. 2001;106:697–708. - PubMed
-
- Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. - PubMed
-
- Suzuki M, Hayashizaki Y. Mouse-centric comparative transcriptomics of protein coding and non-coding RNAs. Bioessays. 2004;26:833–843. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

