The Torso signaling pathway modulates a dual transcriptional switch to regulate tailless expression
- PMID: 19129218
- PMCID: PMC2651784
- DOI: 10.1093/nar/gkn1036
The Torso signaling pathway modulates a dual transcriptional switch to regulate tailless expression
Abstract
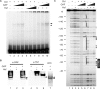
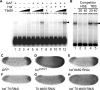
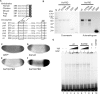
The Torso (Tor) signaling pathway activates tailless (tll) expression by relieving tll repression. None of the repressors identified so far, such as Capicuo, Groucho and Tramtrack69 (Ttk69), bind to the tor response element (tor-RE) or fully elucidate tll repression. In this study, an expanded tll expression pattern was shown in embryos with reduced heat shock factor (hsf) and Trithorax-like (Trl) activities. The GAGA factor, GAF encoded by Trl, bound weakly to the tor-RE, and this binding was enhanced by both Hsf and Ttk69. A similar extent of expansion of tll expression was observed in embryos with simultaneous knockdown of hsf, Trl and ttk69 activities, and in embryos with constitutively active Tor. Hsf is a substrate of mitogen-activated protein kinase and S378 is the major phosphorylation site. Phosphorylation converts Hsf from a repressor to an activator that works with GAF to activate tll expression. In conclusion, the GAF/Hsf/Ttk69 complex binding to the tor-RE remodels local chromatin structure to repress tll expression and the Tor signaling pathway activate tll expression by modulating a dual transcriptional switch.
Figures


 (binding sites for Hsf and GAF are underlined and shadowed, respectively), and a Ttk69 binding site in the even-skipped cis-regulatory region, TCCTCAT
(binding sites for Hsf and GAF are underlined and shadowed, respectively), and a Ttk69 binding site in the even-skipped cis-regulatory region, TCCTCAT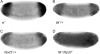
Similar articles
-
A tale of tailless.Dev Neurosci. 2011;33(1):1-13. doi: 10.1159/000321585. Epub 2010 Dec 2. Dev Neurosci. 2011. PMID: 21124006 Free PMC article. Review.
-
Tramtrack69 is required for the early repression of tailless expression.Mech Dev. 2002 Aug;116(1-2):75-83. doi: 10.1016/s0925-4773(02)00143-0. Mech Dev. 2002. PMID: 12128207
-
The torso response element binds GAGA and NTF-1/Elf-1, and regulates tailless by relief of repression.Genes Dev. 1995 Dec 15;9(24):3163-76. doi: 10.1101/gad.9.24.3163. Genes Dev. 1995. PMID: 8543159
-
Drosophila female sterile (1) homeotic is a multifunctional transcriptional regulator that is modulated by Ras signaling.Dev Dyn. 2008 Mar;237(3):554-64. doi: 10.1002/dvdy.21432. Dev Dyn. 2008. PMID: 18264999
-
GAGA factor: a multifunctional pioneering chromatin protein.Cell Mol Life Sci. 2021 May;78(9):4125-4141. doi: 10.1007/s00018-021-03776-z. Epub 2021 Feb 2. Cell Mol Life Sci. 2021. PMID: 33528710 Free PMC article. Review.
Cited by
-
Pits, a protein interacting with Ttk69 and Sin3A, has links to histone deacetylation.Sci Rep. 2016 Sep 13;6:33388. doi: 10.1038/srep33388. Sci Rep. 2016. PMID: 27622813 Free PMC article.
-
Identification of an intronic cis-acting element in the human dopamine transporter gene.Mol Biol Rep. 2012 May;39(5):5393-9. doi: 10.1007/s11033-011-1339-4. Epub 2011 Dec 13. Mol Biol Rep. 2012. PMID: 22160470 Free PMC article.
-
Global repression by tailless during segmentation.Dev Biol. 2024 Jan;505:11-23. doi: 10.1016/j.ydbio.2023.09.014. Epub 2023 Oct 24. Dev Biol. 2024. PMID: 37879494 Free PMC article.
-
OVO positively regulates essential maternal pathways by binding near the transcriptional start sites in the Drosophila female germline.Elife. 2024 Sep 18;13:RP94631. doi: 10.7554/eLife.94631. Elife. 2024. PMID: 39291827 Free PMC article.
-
A tale of tailless.Dev Neurosci. 2011;33(1):1-13. doi: 10.1159/000321585. Epub 2010 Dec 2. Dev Neurosci. 2011. PMID: 21124006 Free PMC article. Review.
References
-
- Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. - PubMed
-
- Hill CS, Treisman R. Transcriptional regulation by extracellular signals: mechanisms and specificity. Cell. 1995;80:199–211. - PubMed
-
- Barolo S, Posakony JW. Three habits of highly effective signaling pathways: principles of transcriptional control by developmental cell signaling. Genes Dev. 2002;16:1167–1181. - PubMed
-
- Sprenger F, Stevens LM, Nusslein-Volhard C. The Drosophila gene torso encodes a putative receptor tyrosine kinase. Nature. 1989;338:478–483. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

