Sensitive fluorescence detection of nucleic acids based on isothermal circular strand-displacement polymerization reaction
- PMID: 19129227
- PMCID: PMC2647313
- DOI: 10.1093/nar/gkn1024
Sensitive fluorescence detection of nucleic acids based on isothermal circular strand-displacement polymerization reaction
Abstract
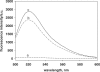
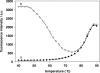
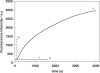
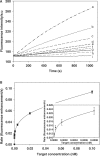
Here we have developed a sensitive DNA amplified detection method based on isothermal strand-displacement polymerization reaction. This method takes advantage of both the hybridization property of DNA and the strand-displacement property of polymerase. Importantly, we demonstrate that our method produces a circular polymerization reaction activated by the target, which essentially allows it to self-detect. Functionally, this DNA system consists of a hairpin fluorescence probe, a short primer and polymerase. Upon recognition and hybridization with the target ssDNA, the stem of the hairpin probe is opened, after which the opened probe anneals with the primer and triggers the polymerization reaction. During this process of the polymerization reaction, a complementary DNA is synthesized and the hybridized target is displaced. Finally, the displaced target recognizes and hybridizes with another probe, triggering the next round of polymerization reaction, reaching a target detection limit of 6.4 x 10(-15) M.
Figures


References
-
- Willner I, Willner B. Biomaterials integrated with electronic elements: enroute to bioelectronics. Trends Biotechnol. 2001;19:222–230. - PubMed
-
- Tom NG, Lars R, Oliver S. Triplex molecular beacons as modular probes for DNA detection. Angew. Chem. Int. Ed. 2007;46:5223–5225. - PubMed
-
- Caruana DJ, Heller A. Enzyme-amplified amperometric detection of hybridization and of a single base pair mutation in an 18-base oligonucleotide on a 7-mm-diameter microelectrode. J. Am. Chem. Soc. 1999;121:769–774.
-
- Patolsky F, Lichtenstein A, Willner I. Detection of single-base DNA mutations by enzyme-amplified electronic transduction. Nat. Biotechnol. 2001;19:253–257. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

