Telomere length measurement by a novel monochrome multiplex quantitative PCR method
- PMID: 19129229
- PMCID: PMC2647324
- DOI: 10.1093/nar/gkn1027
Telomere length measurement by a novel monochrome multiplex quantitative PCR method
Abstract
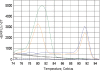
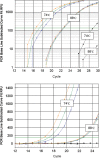
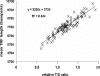
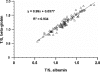
The current quantitative polymerase chain reaction (QPCR) assay of telomere length measures telomere (T) signals in experimental DNA samples in one set of reaction wells, and single copy gene (S) signals in separate wells, in comparison to a reference DNA, to yield relative T/S ratios that are proportional to average telomere length. Multiplexing this assay is desirable, because variation in the amount of DNA pipetted would no longer contribute to variation in T/S, since T and S would be collected within each reaction, from the same input DNA. Multiplexing also increases throughput and lowers costs, since half as many reactions are needed. Here, we present the first multiplexed QPCR method for telomere length measurement. Remarkably, a single fluorescent DNA-intercalating dye is sufficient in this system, because T signals can be collected in early cycles, before S signals rise above baseline, and S signals can be collected at a temperature that fully melts the telomere product, sending its signal to baseline. The correlation of T/S ratios with Terminal Restriction Fragment (TRF) lengths measured by Southern blot was stronger with this monochrome multiplex QPCR method (R(2) = 0.844) than with our original singleplex method (R(2) = 0.677). Multiplex T/S results from independent runs on different days were highly reproducible (R(2) = 0.91).
Figures



References
-
- Kainz P. The PCR plateau phase—towards an understanding of its limitations. Biochim. Biophys. Acta. 2000;1494:23–27. - PubMed
-
- Lawyer FC, Stoffel S, Saiki RK, Chang SY, Landre PA, Abramson RD, Gelfand DH. High-level expression, purification, and enzymatic characterization of full-length Thermus aquaticus DNA polymerase and a truncated form deficient in 5′ to 3′ exonuclease activity. PCR Methods Appl. 1993;2:275–287. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

