Positioning of subdomain IIId and apical loop of domain II of the hepatitis C IRES on the human 40S ribosome
- PMID: 19129232
- PMCID: PMC2651777
- DOI: 10.1093/nar/gkn1026
Positioning of subdomain IIId and apical loop of domain II of the hepatitis C IRES on the human 40S ribosome
Abstract
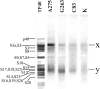
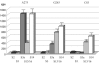
The 5'-untranslated region of the hepatitis C virus (HCV) RNA contains a highly structured motif called IRES (Internal Ribosome Entry Site) responsible for the cap-independent initiation of the viral RNA translation. At first, the IRES binds to the 40S subunit without any initiation factors so that the initiation AUG codon falls into the P site. Here using an original site-directed cross-linking strategy, we identified 40S subunit components neighboring subdomain IIId, which is critical for HCV IRES binding to the subunit, and apical loop of domain II, which was suggested to contact the 40S subunit from data on cryo-electron microscopy of ribosomal complexes containing the HCV IRES. HCV IRES derivatives that bear a photoactivatable group at nucleotide A275 or at G263 in subdomain IIId cross-link to ribosomal proteins S3a, S14 and S16, and HCV IRES derivatized at the C83 in the apex of domain II cross-link to proteins S14 and S16.
Figures


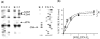


References
-
- Brocard M, Paulous S, Komarova AV, Deveaux V, Kean KM. Evidence that PTB does not stimulate HCV IRES-driven translation. Virus Genes. 2007;35:5–15. - PubMed
-
- Wasley A, Alter MJ. Epidemiology of hepatitis C: geographic differences and temporal trends. Semin. Liver Dis. 2000;20:1–16. - PubMed
-
- Pawlotsky JM. Pathophysiology of hepatitis C virus infection and related liver disease. Trends Microbiol. 2004;12:96–102. - PubMed
-
- Rosenberg S. Recent advances in the molecular biology of hepatitis C virus. J. Mol. Biol. 2001;313:451–464. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

