Essential role of hIST1 in cytokinesis
- PMID: 19129480
- PMCID: PMC2649264
- DOI: 10.1091/mbc.e08-05-0474
Essential role of hIST1 in cytokinesis
Abstract
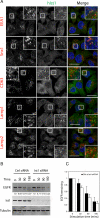
The last steps of multivesicular body (MVB) formation, human immunodeficiency virus (HIV)-1 budding and cytokinesis require a functional endosomal sorting complex required for transport (ESCRT) machinery to facilitate topologically equivalent membrane fission events. Increased sodium tolerance (IST) 1, a new positive modulator of the ESCRT pathway, has been described recently, but an essential function of this highly conserved protein has not been identified. Here, we describe the previously uncharacterized KIAA0174 as the human homologue of IST1 (hIST1), and we report its conserved interaction with VPS4, CHMP1A/B, and LIP5. We also identify a microtubule interacting and transport (MIT) domain interacting motif (MIM) in hIST1 that is necessary for its interaction with VPS4, LIP5 and other MIT domain-containing proteins, namely, MITD1, AMSH, UBPY, and Spastin. Importantly, hIST1 is essential for cytokinesis in mammalian cells but not for HIV-1 budding, thus providing a novel mechanism of functional diversification of the ESCRT machinery. Last, we show that the hIST1 MIM activity is essential for cytokinesis, suggesting possible mechanisms to explain the role of hIST1 in the last step of mammalian cell division.
Figures








References
-
- Agromayor M., Martin-Serrano J. Interaction of AMSH with ESCRT-III and deubiquitination of endosomal cargo. J. Biol. Chem. 2006;281:23083–23091. - PubMed
-
- Azmi I. F., Davies B. A., Xiao J., Babst M., Xu Z., Katzmann D. J. ESCRT-III family members stimulate Vps4 ATPase activity directly or via Vta1. Dev. Cell. 2008;14:50–61. - PubMed
-
- Babst M., Katzmann D. J., Estepa-Sabal E. J., Meerloo T., Emr S. D. Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev. Cell. 2002a;3:271–282. - PubMed
-
- Babst M., Katzmann D. J., Snyder W. B., Wendland B., Emr S. D. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell. 2002b;3:283–289. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

