Nkx2-5 transactivates the Ets-related protein 71 gene and specifies an endothelial/endocardial fate in the developing embryo
- PMID: 19129488
- PMCID: PMC2630085
- DOI: 10.1073/pnas.0807583106
Nkx2-5 transactivates the Ets-related protein 71 gene and specifies an endothelial/endocardial fate in the developing embryo
Abstract
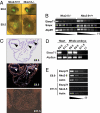
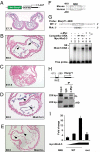
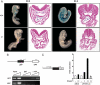
Recent studies support the existence of a common progenitor for the cardiac and endothelial cell lineages, but the underlying transcriptional networks responsible for specification of these cell fates remain unclear. Here we demonstrated that Ets-related protein 71 (Etsrp71), a newly discovered ETS family transcription factor, was a novel downstream target of the homeodomain protein, Nkx2-5. Using genetic mouse models and molecular biological techniques, we demonstrated that Nkx2-5 binds to an evolutionarily conserved Nkx2-5 response element in the Etsrp71 promoter and induces the Etsrp71 gene expression in vitro and in vivo. Etsrp71 was transiently expressed in the endocardium/endothelium of the developing embryo (E7.75-E9.5) and was extinguished during the latter stages of development. Using a gene disruption strategy, we found that Etsrp71 mutant embryos lacked endocardial/endothelial lineages and were nonviable. Moreover, using transgenic technologies and transcriptional and chromatin immunoprecipitation (ChIP) assays, we further established that Tie2 is a direct downstream target of Etsrp71. Collectively, our results uncover a novel functional role for Nkx2-5 and define a transcriptional network that specifies an endocardial/endothelial fate in the developing heart and embryo.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Olson EN, Srivastava D. Molecular pathways controlling heart development. Science. 1996;272:671–676. - PubMed
-
- Fishman MC, Chien KR. Fashioning the vertebrate heart: earliest embryonic decisions. Development. 1997;124:2099–2117. - PubMed
-
- Kattman SJ, Huber TL, Keller GM. Multipotent flk-1+ cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial, and vascular smooth muscle lineages. Dev Cell. 2006;11:723–732. - PubMed
-
- Wu SM, et al. Developmental origin of a bipotential myocardial and smooth muscle cell precursor in the mammalian heart. Cell. 2006;127:1137–1150. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

