Dynamic nuclear organization of constitutive heterochromatin during fetal male germ cell development in mice
- PMID: 19129513
- PMCID: PMC2804833
- DOI: 10.1095/biolreprod.108.072603
Dynamic nuclear organization of constitutive heterochromatin during fetal male germ cell development in mice
Abstract
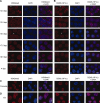
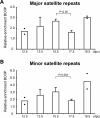
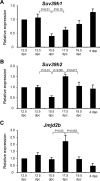
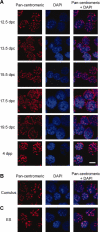
In mice, male germ cells enter mitotic arrest beginning at 13.5 days postcoitum (dpc), and remain suspended in the G(0)/G(1) cell cycle stage until after birth. During this period, male germ cells undergo extensive epigenetic reprogramming, which is essential for their subsequent function as male gametes. A global reorganization and spatial clustering of constitutive heterochromatin has been implicated in epigenetic plasticity during cellular differentiation. Here, we have studied the dynamics of heterochromatin in fetal (12.5-19.5 dpc) and neonatal (4 days postpartum) male germ cells. We monitored constitutive heterochromatin-specific markers, and observed changes in the association of histone H3 trimethylation of lysine 9 (H3K9me3), binding of heterochromatin protein 1, and patterns of 4',6-diamino-2-phenylindole staining in pericentric regions of chromosomes, along with a coincident loss of chromocenters in fetal prospermatogonia during mitotic arrest. We also observed a transient loss of H3K9me3 associated with major and minor satellite repeat sequences, plus inactivation of histone methyltransferases (Suv39h1 and Suv39h2), and transient activation of histone demethylase (Jmjd2b) in these same cells. These epigenetic changes were correlated with relocation of centromeric regions toward the nuclear periphery in prospermatogonia during mitotic arrest. Taken together, these results show that constitutive heterochromatin undergoes dramatic reorganization during prespermatogenesis. We suggest that these dynamic changes in heterochromatin contribute to normal epigenetic reprogramming of the paternal genome in fetal prospermatogonia suspended in the G(0)/G(1) stage, and that this also represents an epigenomic state that is particularly amenable to reprogramming.
Figures

References
-
- Ohinata Y, Payer B, O'Carroll D, Ancelin K, Ono Y, Sano M, Barton SC, Obukhanych T, Nussenzweig M, Tarakhovsky A, Saitou M, Surani MA.Blimp1 is a critical determinant of the germ cell lineage in mice. Nature 2005; 436: 207–213. - PubMed
-
- McCarrey JR.Development of the germ cell. Desjardins C, Ewing LL.Cell and Molecular Biology of the Testis New York:Oxford University Press;1993: 58–89.
-
- McLaren A.Primordial germ cells in the mouse. Dev Biol 2003; 262: 1–15. - PubMed
-
- Koopman P, Münsterberg A, Capel B, Vivian N, Lovell-Badge R.Expression of a candidate sex-determining gene during mouse testis differentiation. Nature 1990; 348: 450–452. - PubMed
-
- Wilhelm D, Koopman P.The makings of maleness: towards an integrated view of male sexual development. Nat Rev Genet 2006; 7: 620–631. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

