Identification and characterization of a novel nuclear protein complex involved in nuclear hormone receptor-mediated gene regulation
- PMID: 19131338
- PMCID: PMC2658049
- DOI: 10.1074/jbc.M805872200
Identification and characterization of a novel nuclear protein complex involved in nuclear hormone receptor-mediated gene regulation
Abstract
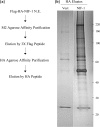
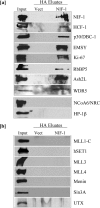
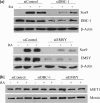
NRC/NCoA6 plays an important role in mediating the effects of ligand-bound nuclear hormone receptors as well as other transcription factors. NRC interacting factor 1 (NIF-1) was cloned as a novel factor that interacts in vivo with NRC. Although NIF-1 does not directly interact with nuclear hormone receptors, it enhances activation by nuclear hormone receptors presumably through its interaction with NRC. To further understand the cellular and biological function of NIF-1, we identified NIF-1-associated proteins by in-solution proteolysis followed by mass spectrometry. The identified components revealed factors involved in histone methylation and cell cycle control and include Ash2L, RbBP5, WDR5, HCF-1, DBC-1, and EMSY. Although the NIF-1 complex contains Ash2L, RbBP5, and WDR5, suggesting that the complex might methylate histone H3-Lys-4, we found that the complex contains a H3 methyltransferase activity that modifies a residue other than H3-Lys-4. The identified components form at least two distinctly sized NIF-1 complexes. DBC-1 and EMSY were identified as integral components of an NIF-1 complex of approximately 1.5 MDa and were found to play an important role in the regulation of nuclear receptor-mediated transcription. Stimulation of the Sox9 and HoxA1 genes by retinoic acid receptor-alpha was found to require both DBC-1 and EMSY in addition to NIF-1 for maximal transcriptional activation. Interestingly, NRC was not identified as a component of the NIF-1 complex, suggesting that NIF-1 and NRC do not exist as stable in vitro purified complexes, although the separate NIF-1 and NRC complexes appear to functionally interact in the cell.
Figures

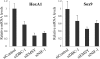



References
-
- McKenna, N. J., and O'Malley, B. W. (2002) Cell 108 465–474 - PubMed
-
- Lonard, D. M., Lanz, R. B., and O'Malley, B. W. (2007) Endocr. Rev. 28 575–587 - PubMed
-
- Lonard, D. M., and O'Malley, B. W. (2007) Mol. Cell 27 691–700 - PubMed
-
- Demarest, S. J., Martinez-Yamout, M., Chung, J., Chen, H., Xu, W., Dyson, H. J., Evans, R. M., and Wright, P. E. (2002) Nature 415 549–553 - PubMed
-
- Xu, J., and Li, Q. (2003) Mol. Endocrinol. 17 1681–1692 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

