Remodelling of VipA/VipB tubules by ClpV-mediated threading is crucial for type VI protein secretion
- PMID: 19131969
- PMCID: PMC2646146
- DOI: 10.1038/emboj.2008.269
Remodelling of VipA/VipB tubules by ClpV-mediated threading is crucial for type VI protein secretion
Abstract
The recently identified type VI secretion systems (T6SS) have a crucial function in the virulence of various proteobacteria, including the human pathogen Vibrio cholerae. T6SS are encoded by a conserved gene cluster comprising approximately 15 open reading frames, mediating the appearance of Hcp and VgrG proteins in cell culture supernatants. Here, we analysed the function of the V. cholerae T6SS member ClpV, a specialized AAA+ protein. ClpV is crucial for a functional T6SS and interacts through its N-terminal domain with the VipA/VipB complex that is composed of two conserved and essential members of T6SS. Transferring ClpV substrate specificity to a distinct AAA+ protein involved in proteolysis caused degradation of VipA but not Hcp or VgrG2, suggesting that VipA rather than Hcp/VgrG2 functions as a primary ClpV substrate. Strikingly, VipA/VipB form tubular, cogwheel-like structures that are converted by a threading activity of ClpV into small complexes. ClpV-mediated remodelling of VipA/VipB tubules represents a crucial step in T6S, illuminating an unexpected role of an ATPase component in protein secretion.
Figures


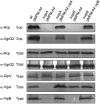

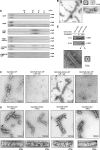

Comment in
-
The type VI secretion system: a tubular story.EMBO J. 2009 Feb 18;28(4):309-10. doi: 10.1038/emboj.2008.301. EMBO J. 2009. PMID: 19225443 Free PMC article.
References
-
- Akeda Y, Galan JE (2005) Chaperone release and unfolding of substrates in type III secretion. Nature 437: 911–915 - PubMed
-
- Bladergroen MR, Badelt K, Spaink HP (2003) Infection-blocking genes of a symbiotic Rhizobium leguminosarum strain that are involved in temperature-dependent protein secretion. Mol Plant Microbe Interact 16: 53–64 - PubMed
-
- Das S, Chaudhuri K (2003) Identification of a unique IAHP (IcmF associated homologous proteins) cluster in Vibrio cholerae and other proteobacteria through in silico analysis. In Silico Biol 3: 287–300 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

