Why is the correlation between gene importance and gene evolutionary rate so weak?
- PMID: 19132081
- PMCID: PMC2605560
- DOI: 10.1371/journal.pgen.1000329
Why is the correlation between gene importance and gene evolutionary rate so weak?
Abstract
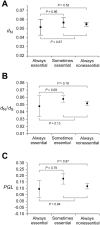
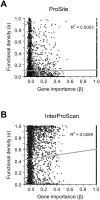
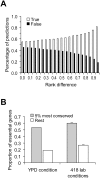
One of the few commonly believed principles of molecular evolution is that functionally more important genes (or DNA sequences) evolve more slowly than less important ones. This principle is widely used by molecular biologists in daily practice. However, recent genomic analysis of a diverse array of organisms found only weak, negative correlations between the evolutionary rate of a gene and its functional importance, typically measured under a single benign lab condition. A frequently suggested cause of the above finding is that gene importance determined in the lab differs from that in an organism's natural environment. Here, we test this hypothesis in yeast using gene importance values experimentally determined in 418 lab conditions or computationally predicted for 10,000 nutritional conditions. In no single condition or combination of conditions did we find a much stronger negative correlation, which is explainable by our subsequent finding that always-essential (enzyme) genes do not evolve significantly more slowly than sometimes-essential or always-nonessential ones. Furthermore, we verified that functional density, approximated by the fraction of amino acid sites within protein domains, is uncorrelated with gene importance. Thus, neither the lab-nature mismatch nor a potentially biased among-gene distribution of functional density explains the observed weakness of the correlation between gene importance and evolutionary rate. We conclude that the weakness is factual, rather than artifactual. In addition to being weakened by population genetic reasons, the correlation is likely to have been further weakened by the presence of multiple nontrivial rate determinants that are independent from gene importance. These findings notwithstanding, we show that the principle of slower evolution of more important genes does have some predictive power when genes with vastly different evolutionary rates are compared, explaining why the principle can be practically useful despite the weakness of the correlation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Karp G. Cell and Molecular Biology. Hoboken, New Jersey: John Wiley & Sons, Inc; 2008.
-
- Liao BY, Scott NM, Zhang J. Impacts of gene essentiality, expression pattern, and gene compactness on the evolutionary rate of mammalian proteins. Mol Biol Evol. 2006;23:2072–2080. - PubMed
-
- Rocha EP, Danchin A. An analysis of determinants of amino acids substitution rates in bacterial proteins. Mol Biol Evol. 2004;21:108–116. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

