Role of intraspecies recombination in the spread of pathogenicity islands within the Escherichia coli species
- PMID: 19132082
- PMCID: PMC2606025
- DOI: 10.1371/journal.ppat.1000257
Role of intraspecies recombination in the spread of pathogenicity islands within the Escherichia coli species
Abstract
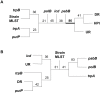
Horizontal gene transfer is a key step in the evolution of bacterial pathogens. Besides phages and plasmids, pathogenicity islands (PAIs) are subjected to horizontal transfer. The transfer mechanisms of PAIs within a certain bacterial species or between different species are still not well understood. This study is focused on the High-Pathogenicity Island (HPI), which is a PAI widely spread among extraintestinal pathogenic Escherichia coli and serves as a model for horizontal transfer of PAIs in general. We applied a phylogenetic approach using multilocus sequence typing on HPI-positive and -negative natural E. coli isolates representative of the species diversity to infer the mechanism of horizontal HPI transfer within the E. coli species. In each strain, the partial nucleotide sequences of 6 HPI-encoded genes and 6 housekeeping genes of the genomic backbone, as well as DNA fragments immediately upstream and downstream of the HPI were compared. This revealed that the HPI is not solely vertically transmitted, but that recombination of large DNA fragments beyond the HPI plays a major role in the spread of the HPI within E. coli species. In support of the results of the phylogenetic analyses, we experimentally demonstrated that HPI can be transferred between different E. coli strains by F-plasmid mediated mobilization. Sequencing of the chromosomal DNA regions immediately upstream and downstream of the HPI in the recipient strain indicated that the HPI was transferred and integrated together with HPI-flanking DNA regions of the donor strain. The results of this study demonstrate for the first time that conjugative transfer and homologous DNA recombination play a major role in horizontal transfer of a pathogenicity island within the species E. coli.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

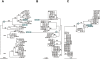
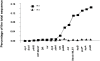

References
-
- Arber W. Evolution of prokaryotic genomes. Gene. 1993;135:49–56. - PubMed
-
- Chen I, Dubnau D. DNA uptake during bacterial transformation. Nat Rev Microbiol. 2004;2:241–249. - PubMed
-
- Hacker J, Kaper JB. Pathogenicity islands and the evolution of microbes. Annu Rev Microbiol. 2000;54:641–679. - PubMed
-
- Kaper JB, McDaniel TK, Jarvis KG, Gomez-Duarte O. Genetics of virulence of enteropathogenic E. coli. Adv Exp Med Biol. 1997;412:279–287. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

