The ABC transporter MsbA interacts with lipid A and amphipathic drugs at different sites
- PMID: 19132955
- PMCID: PMC2662489
- DOI: 10.1042/BJ20081364
The ABC transporter MsbA interacts with lipid A and amphipathic drugs at different sites
Abstract
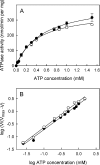
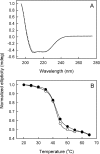
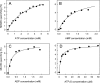
MsbA is an essential ABC (ATP-binding cassette) transporter involved in lipid A transport across the cytoplasmic membrane of Gram-negative bacteria. The protein has also been linked to efflux of amphipathic drugs. Purified wild-type MsbA was labelled stoichiometrically with the fluorescent probe MIANS [2-(4'-maleimidylanilino)naphthalene-6-sulfonic acid] on C315, which is located within the intracellular domain connecting transmembrane helix 6 and the nucleotide-binding domain. MsbA-MIANS displayed high ATPase activity, and its folding and stability were unchanged. The initial rate of MsbA labelling by MIANS was reduced in the presence of amphipathic drugs, suggesting that binding of these compounds alters the protein conformation. The fluorescence of MsbA-MIANS was saturably quenched by nucleotides, lipid A and various drugs, and estimates of the Kd values for binding fell in the range of 0.35-10 microM. Lipid A and daunorubicin were able to bind to MsbA-MIANS simultaneously, implying that they occupy different binding sites. The effects of nucleotide and lipid A/daunorubicin binding were additive, and binding was not ordered. The Kd of MsbA for binding lipid A was substantially decreased when the daunorubicin binding site was occupied first, and prior binding of nucleotide also modulated lipid A binding affinity. These results indicate that MsbA contains two substrate-binding sites that communicate with both the nucleotide-binding domain and with each other. One is a high affinity binding site for the physiological substrate, lipid A, and the other site interacts with drugs with comparable affinity. Thus MsbA may function as both a lipid flippase and a multidrug transporter.
Figures




References
-
- Dassa E., Bouige P. The ABC of ABCs: a phylogenetic and functional classification of ABC systems in living organisms. Res. Microbiol. 2001;152:211–229. - PubMed
-
- Davidson A. L., Chen J. ATP-binding cassette transporters in bacteria. Annu. Rev. Biochem. 2004;73:241–268. - PubMed
-
- Higgins C. F. ABC transporters: physiology, structure and mechanism – an overview. Res. Microbiol. 2001;152:205–210. - PubMed
-
- Doerrler W. T., Reedy M. C., Raetz C. R. An Escherichia coli mutant defective in lipid export. J. Biol. Chem. 2001;276:11461–11464. - PubMed
-
- Doerrler W. T., Gibbons H. S., Raetz C. R. MsbA-dependent translocation of lipids across the inner membrane of Escherichia coli. J. Biol. Chem. 2004;279:45102–45109. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

