A global benchmark study using affinity-based biosensors
- PMID: 19133223
- PMCID: PMC3793259
- DOI: 10.1016/j.ab.2008.11.021
A global benchmark study using affinity-based biosensors
Abstract
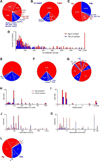
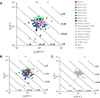
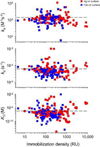
To explore the variability in biosensor studies, 150 participants from 20 countries were given the same protein samples and asked to determine kinetic rate constants for the interaction. We chose a protein system that was amenable to analysis using different biosensor platforms as well as by users of different expertise levels. The two proteins (a 50-kDa Fab and a 60-kDa glutathione S-transferase [GST] antigen) form a relatively high-affinity complex, so participants needed to optimize several experimental parameters, including ligand immobilization and regeneration conditions as well as analyte concentrations and injection/dissociation times. Although most participants collected binding responses that could be fit to yield kinetic parameters, the quality of a few data sets could have been improved by optimizing the assay design. Once these outliers were removed, the average reported affinity across the remaining panel of participants was 620 pM with a standard deviation of 980 pM. These results demonstrate that when this biosensor assay was designed and executed appropriately, the reported rate constants were consistent, and independent of which protein was immobilized and which biosensor was used.
Figures

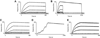
















References
-
- Rich RL, Myszka DG. Survey of the year 2005 commercial optical biosensor literature. J. Mol. Recognit. 2006;19:478–534. - PubMed
-
- Rich RL, Myszka DG. Survey of the year 2006 commercial optical biosensor literature. J. Mol. Recognit. 2007;20:300–366. - PubMed
-
- Rich RL, Myszka DG. Survey of the year 2007 commercial optical biosensor literature. J. Mol. Recognit. 2008;21:355–400. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

