The G-alpha protein GNA3 of Hypocrea jecorina (Anamorph Trichoderma reesei) regulates cellulase gene expression in the presence of light
- PMID: 19136572
- PMCID: PMC2653238
- DOI: 10.1128/EC.00256-08
The G-alpha protein GNA3 of Hypocrea jecorina (Anamorph Trichoderma reesei) regulates cellulase gene expression in the presence of light
Abstract
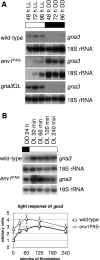
Although the enzymes enabling Hypocrea jecorina (anamorph Trichoderma reesei) to degrade the insoluble substrate cellulose have been investigated in some detail, little is still known about the mechanism by which cellulose signals its presence to the fungus. In order to investigate the possible role of a G-protein/cyclic AMP signaling pathway, the gene encoding GNA3, which belongs to the adenylate cyclase-activating class III of G-alpha subunits, was cloned. gna3 is clustered in tandem with the mitogen-activated protein kinase gene tmk3 and the glycogen phosphorylase gene gph1. The gna3 transcript is upregulated in the presence of light and is almost absent in the dark. A strain bearing a constitutively activated version of GNA3 (gna3QL) exhibits strongly increased cellulase transcription in the presence of the inducer cellulose and in the presence of light, whereas a gna3 antisense strain showed delayed cellulase transcription under this condition. However, the gna3QL mutant strain was unable to form cellulases in the absence of cellulose. The necessity of light for stimulation of cellulase transcription by GNA3 could not be overcome in a mutant which expressed gna3 under control of the constitutive gpd1 promoter also in darkness. We conclude that the previously reported stimulation of cellulase gene transcription by light, but not the direct transmission of the cellulose signal, involves the function and activation of GNA3. The upregulation of gna3 by light is influenced by the light modulator ENVOY, but GNA3 itself has no effect on transcription of the light regulator genes blr1, blr2, and env1. Our data for the first time imply an involvement of a G-alpha subunit in a light-dependent signaling event in fungi.
Figures





References
-
- Amaar, Y. G., and M. M. Moore. 1998. Mapping of the nitrate-assimilation gene cluster (crnA-niiA-niaD) and characterization of the nitrite reductase gene (niiA) in the opportunistic fungal pathogen Aspergillus fumigatus. Curr. Genet. 33206-215. - PubMed
-
- Aro, N., T. Pakula, and M. Penttila. 2005. Transcriptional regulation of plant cell wall degradation by filamentous fungi. FEMS Microbiol. Rev. 29719-739. - PubMed
-
- Aro, N., A. Saloheimo, M. Ilmen, and M. Penttila. 2001. ACEII, a novel transcriptional activator involved in regulation of cellulase and xylanase genes of Trichoderma reesei. J. Biol. Chem. 27624309-24314. - PubMed
-
- Berman, D. M., T. M. Wilkie, and A. G. Gilman. 1996. GAIP and RGS4 are GTPase-activating proteins for the Gi subfamily of G protein alpha subunits. Cell 86445-452. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

