Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression
- PMID: 19136627
- PMCID: PMC2632171
- DOI: 10.1101/gad.502409
Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression
Abstract
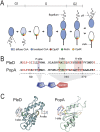
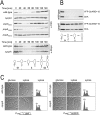
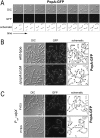
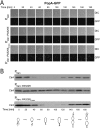
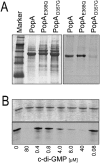
Second messengers control a wide range of important cellular functions in eukaryotes and prokaryotes. Here we show that cyclic di-GMP, a global bacterial second messenger, promotes cell cycle progression in Caulobacter crescentus by mediating the specific degradation of the replication initiation inhibitor CtrA. During the G1-to-S-phase transition, both CtrA and its cognate protease ClpXP dynamically localize to the old cell pole, where CtrA is rapidly degraded. Sequestration of CtrA to the cell pole depends on PopA, a newly identified cyclic di-GMP effector protein. PopA itself localizes to the cell pole and directs CtrA to this subcellular site via the direct interaction with a mediator protein, RcdA. We present evidence that c-di-GMP regulates CtrA degradation during the cell cycle by controlling the dynamic sequestration of the PopA recruitment factor to the cell pole. Furthermore, we show that cell cycle timing of CtrA degradation relies on converging pathways responsible for substrate and protease localization to the old cell pole. This is the first report that links cyclic di-GMP to protein dynamics and cell cycle control in bacteria.
Figures



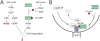
References
-
- Aldridge P., Paul R., Goymer P., Rainey P., Jenal U. Role of the GGDEF regulator PleD in polar development of Caulobacter crescentus . Mol. Microbiol. 2003;47:1695–1708. - PubMed
-
- Biondi E.G., Reisinger S.J., Skerker J.M., Arif M., Perchuk B.S., Ryan K.R., Laub M.T. Regulation of the bacterial cell cycle by an integrated genetic circuit. Nature. 2006;444:899–904. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
