PTMap--a sequence alignment software for unrestricted, accurate, and full-spectrum identification of post-translational modification sites
- PMID: 19136633
- PMCID: PMC2630079
- DOI: 10.1073/pnas.0811739106
PTMap--a sequence alignment software for unrestricted, accurate, and full-spectrum identification of post-translational modification sites
Abstract
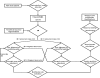
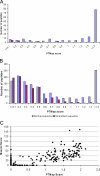
We present sequence alignment software, called PTMap, for the accurate identification of full-spectrum protein post-translational modifications (PTMs) and polymorphisms. The software incorporates several features to improve searching speed and accuracy, including peak selection, adjustment of inaccurate mass shifts, and precise localization of PTM sites. PTMap also automates rules, based mainly on unmatched peaks, for manual verification of identified peptides. To evaluate the quality of sequence alignment, we developed a scoring system that takes into account both matched and unmatched peaks in the mass spectrum. Incorporation of these features dramatically increased both accuracy and sensitivity of the peptide- and PTM-identifications. To our knowledge, PTMap is the first algorithm that emphasizes unmatched peaks to eliminate false positives. The superior performance and reliability of PTMap were demonstrated by confident identification of PTMs on 156 peptides from four proteins and validated by MS/MS of the synthetic peptides. Our results demonstrate that PTMap is a powerful algorithm capable of identification of all possible protein PTMs with high confidence.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Witze ES, Old WM, Resing KA, Ahn NG. Mapping protein post-translational modifications with mass spectrometry. Nat Methods. 2007;4:798–806. - PubMed
-
- Jensen ON. Modification-specific proteomics: Characterization of post-translational modifications by mass spectrometry. Curr Opin Chem Biol. 2004;8:33–41. - PubMed
-
- Sadygov RG, Cociorva D, Yates JR., 3rd Large-scale database searching using tandem mass spectra: Looking up the answer in the back of the book. Nat Methods. 2004;1:195–202. - PubMed
-
- Eng JK, McCormack AL, Yates JR., 3rd An Approach to Correlate Tandem Mass Spectral Data of Peptides with Amino Acid Sequences in a Protein Database. J Am Soc Mass Spectrom. 1994;5:976–989. - PubMed
-
- Yates JR, 3rd, Eng JK, McCormack AL. Mining genomes: Correlating tandem mass spectra of modified and unmodified peptides to sequences in nucleotide databases. Anal Chem. 1995;67:3202–3210. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

