Rearrangement of actin cytoskeleton mediates invasion of Lotus japonicus roots by Mesorhizobium loti
- PMID: 19136645
- PMCID: PMC2648097
- DOI: 10.1105/tpc.108.063693
Rearrangement of actin cytoskeleton mediates invasion of Lotus japonicus roots by Mesorhizobium loti
Abstract
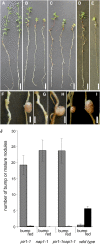
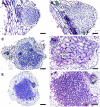
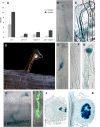
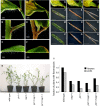
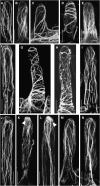
Infection thread-dependent invasion of legume roots by rhizobia leads to internalization of bacteria into the plant cells, which is one of the salient features of root nodule symbiosis. We found that two genes, Nap1 (for Nck-associated protein 1) and Pir1 (for 121F-specific p53 inducible RNA), involved in actin rearrangements were essential for infection thread formation and colonization of Lotus japonicus roots by its natural microsymbiont, Mesorhizobium loti. nap1 and pir1 mutants developed an excess of uncolonized nodule primordia, indicating that these two genes were not essential for the initiation of nodule organogenesis per se. However, both the formation and subsequent progression of infection threads into the root cortex were significantly impaired in these mutants. We demonstrate that these infection defects were due to disturbed actin cytoskeleton organization. Short root hairs of the mutants had mostly transverse or web-like actin filaments, while bundles of actin filaments in wild-type root hairs were predominantly longitudinal. Corroborating these observations, temporal and spatial differences in actin filament organization between wild-type and mutant root hairs were also observed after Nod factor treatment, while calcium influx and spiking appeared unperturbed. Together with various effects on plant growth and seed formation, the nap1 and pir1 alleles also conferred a characteristic distorted trichome phenotype, suggesting a more general role for Nap1 and Pir1 in processes establishing cell polarity or polar growth in L. japonicus.
Figures




Similar articles
-
Lotus japonicus ARPC1 is required for rhizobial infection.Plant Physiol. 2012 Oct;160(2):917-28. doi: 10.1104/pp.112.202572. Epub 2012 Aug 3. Plant Physiol. 2012. PMID: 22864583 Free PMC article.
-
micro RNA 172 (miR172) signals epidermal infection and is expressed in cells primed for bacterial invasion in Lotus japonicus roots and nodules.New Phytol. 2015 Oct;208(1):241-56. doi: 10.1111/nph.13445. Epub 2015 May 13. New Phytol. 2015. PMID: 25967282
-
SCARN a Novel Class of SCAR Protein That Is Required for Root-Hair Infection during Legume Nodulation.PLoS Genet. 2015 Oct 30;11(10):e1005623. doi: 10.1371/journal.pgen.1005623. eCollection 2015 Oct. PLoS Genet. 2015. PMID: 26517270 Free PMC article.
-
Invasion of Lotus japonicus root hairless 1 by Mesorhizobium loti involves the nodulation factor-dependent induction of root hairs.Plant Physiol. 2005 Apr;137(4):1331-44. doi: 10.1104/pp.104.057513. Epub 2005 Mar 18. Plant Physiol. 2005. PMID: 15778455 Free PMC article.
-
Inside out: root cortex-localized LHK1 cytokinin receptor limits epidermal infection of Lotus japonicus roots by Mesorhizobium loti.New Phytol. 2019 May;222(3):1523-1537. doi: 10.1111/nph.15683. Epub 2019 Feb 14. New Phytol. 2019. PMID: 30636324
Cited by
-
SPIKE1 Activates the GTPase ROP6 to Guide the Polarized Growth of Infection Threads in Lotus japonicus.Plant Cell. 2020 Dec;32(12):3774-3791. doi: 10.1105/tpc.20.00109. Epub 2020 Oct 6. Plant Cell. 2020. PMID: 33023954 Free PMC article.
-
Deep Sequencing of the Medicago truncatula Root Transcriptome Reveals a Massive and Early Interaction between Nodulation Factor and Ethylene Signals.Plant Physiol. 2015 Sep;169(1):233-65. doi: 10.1104/pp.15.00350. Epub 2015 Jul 14. Plant Physiol. 2015. PMID: 26175514 Free PMC article.
-
The association of the Arabidopsis actin-related protein2/3 complex with cell membranes is linked to its assembly status but not its activation.Plant Physiol. 2009 Dec;151(4):2095-109. doi: 10.1104/pp.109.143859. Epub 2009 Oct 2. Plant Physiol. 2009. PMID: 19801398 Free PMC article.
-
Innovation and appropriation in mycorrhizal and rhizobial Symbioses.Plant Cell. 2022 Apr 26;34(5):1573-1599. doi: 10.1093/plcell/koac039. Plant Cell. 2022. PMID: 35157080 Free PMC article.
-
Visualization of the Crossroads between a Nascent Infection Thread and the First Cell Division Event in Phaseolus vulgaris Nodulation.Int J Mol Sci. 2022 May 9;23(9):5267. doi: 10.3390/ijms23095267. Int J Mol Sci. 2022. PMID: 35563659 Free PMC article.
References
-
- Allen, N.S., Bennett, M.N., Cox, D.N., Shipley, A., Ehrhardt, D.W., and Long, S.R. (1994). Effects of Nod factors on alfalfa root hair Ca++ and H+ currents and on cytoskeletal behavior. In Advances in Molecular Genetics of Plant-Microbe Interactions, Vol. 3. M.J. Daniels, J.A. Downie, and A.E. Osbourn, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 107–113.
-
- Ané, J.-M., et al. (2004). Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 303 1364–1367. - PubMed
-
- Bannigan, A., and Baskin, T.I. (2005). Directional cell expansion – Turning toward actin. Curr. Opin. Plant Biol. 8 619–624. - PubMed
-
- Basu, D., El-Din El-Assl, S., Le, J., Mallery, E.L., and Szymanski, D.B. (2004). Interchangeable functions of Arabidopsis PIROGI and the human WAVE complex subunit SRA1 during leaf epidermal development. Development 131 4345–4355. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

