Specific regulation of mRNA cap methylation by the c-Myc and E2F1 transcription factors
- PMID: 19137018
- PMCID: PMC2652438
- DOI: 10.1038/onc.2008.463
Specific regulation of mRNA cap methylation by the c-Myc and E2F1 transcription factors
Abstract
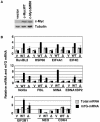
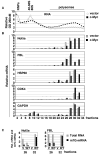
Methylation of the mRNA 5' guanosine cap is essential for efficient gene expression. The 5' methyl cap binds to eIF4E, which is the first step in the recruitment of mRNA to the 40S ribosomal subunit. To investigate whether mRNA cap methylation is regulated in a gene-specific manner, we established a method to detect the relative level of cap methylation on specific mRNAs. We found that two transcription factors, c-Myc and E2F1, induce cap methylation of their transcriptional target genes, and therefore, c-Myc and E2F1 upregulate gene expression by simultaneously inducing transcription and promoting translation. c-Myc-induced cap methylation is greater than transcriptional induction for the majority of its target genes, indicating that this is a major mechanism by which Myc regulates gene expression.
Figures


References
-
- Bentley DL. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol. 2005;17:251–6. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

